Method and system for filtering sequence segments in short-sequence assembly
A filtering method and a technology of sequence fragments, which are applied in the field of genetic engineering, can solve problems such as the inability to analyze gene sequences, consume huge memory, and bubble type errors, and achieve the effects of small errors, reduced short string sets, and improved performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0041] In order to enable those skilled in the art to better understand the technical solutions of the present application, the technical solutions in the embodiments of the present application will be clearly and completely described below in conjunction with the drawings in the embodiments of the present application. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
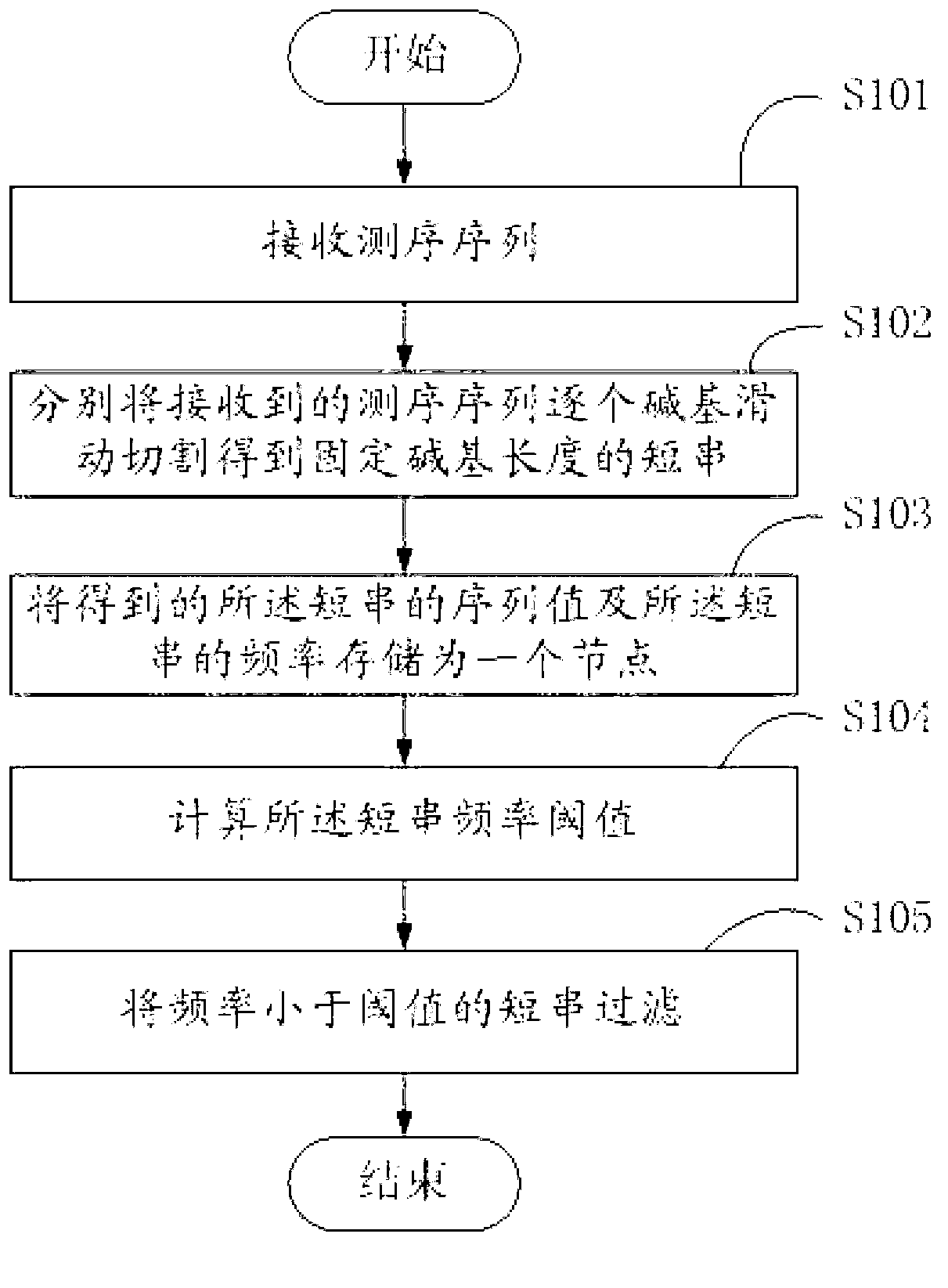
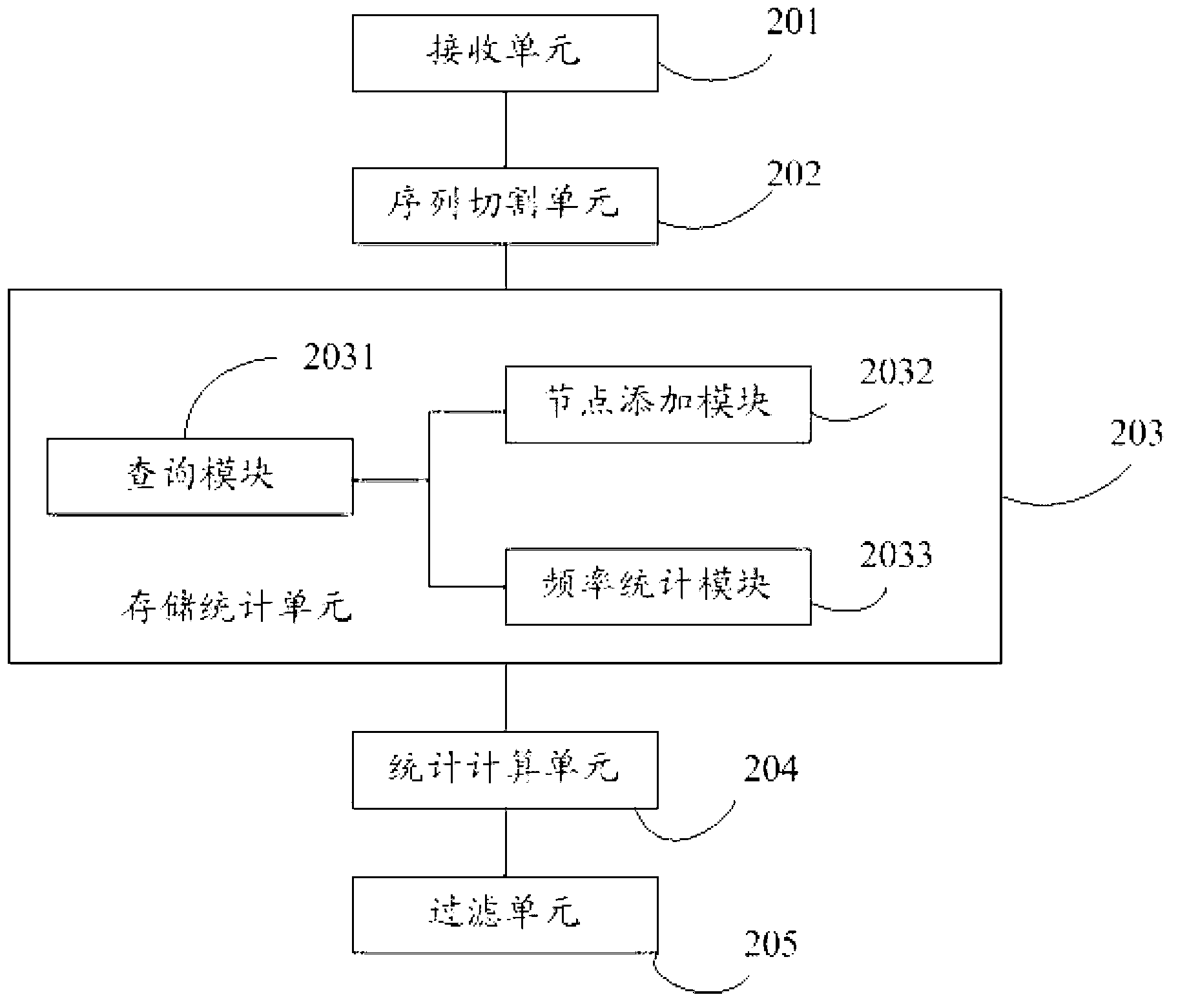
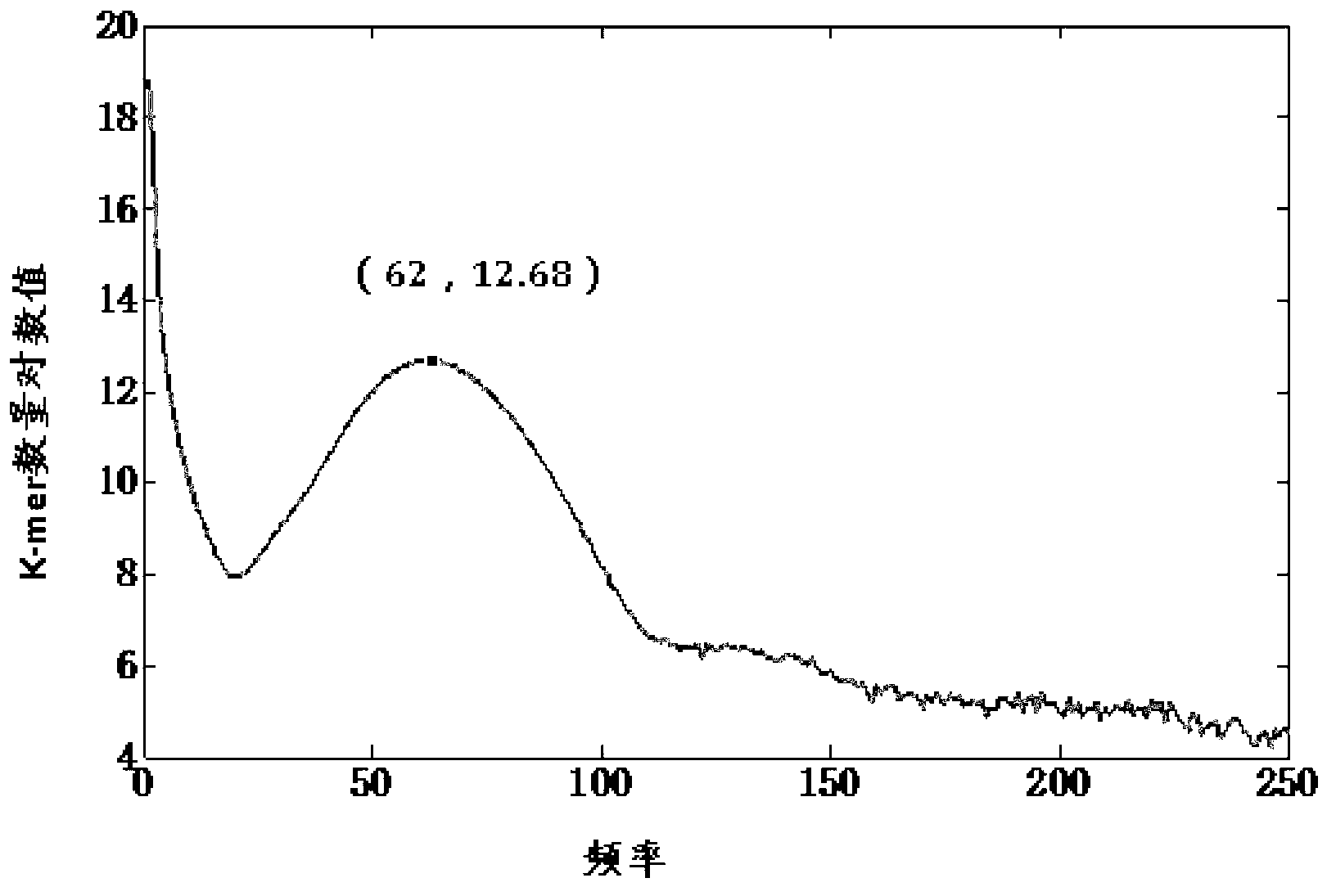
[0042] In the embodiment of the present invention, short strings (k-mer) of fixed base length are obtained by sliding and cutting the received sequencing sequence base by base, and the sequence values of the obtained short strings are stored, and the obtained For each occurrence frequency of the short strings, draw a frequency statistical graph of the short strings, calculate the short string frequency threshold, and filter the short strings whose frequency is less than the threshold.
[0043] figure 1 Show...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com