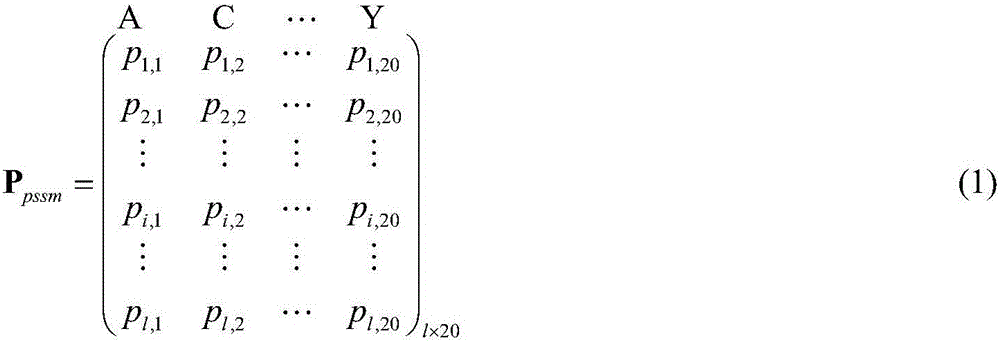Protein-vitamin binding site prediction method based on subspace fusion
A technology for binding sites and prediction methods, which can be applied in the fields of instrumentation, computing, electrical digital data processing, etc., and can solve problems such as poor interpretability and large gaps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0085] In order to better understand the technical content of the present invention, specific embodiments are given together with the attached drawings for description as follows.
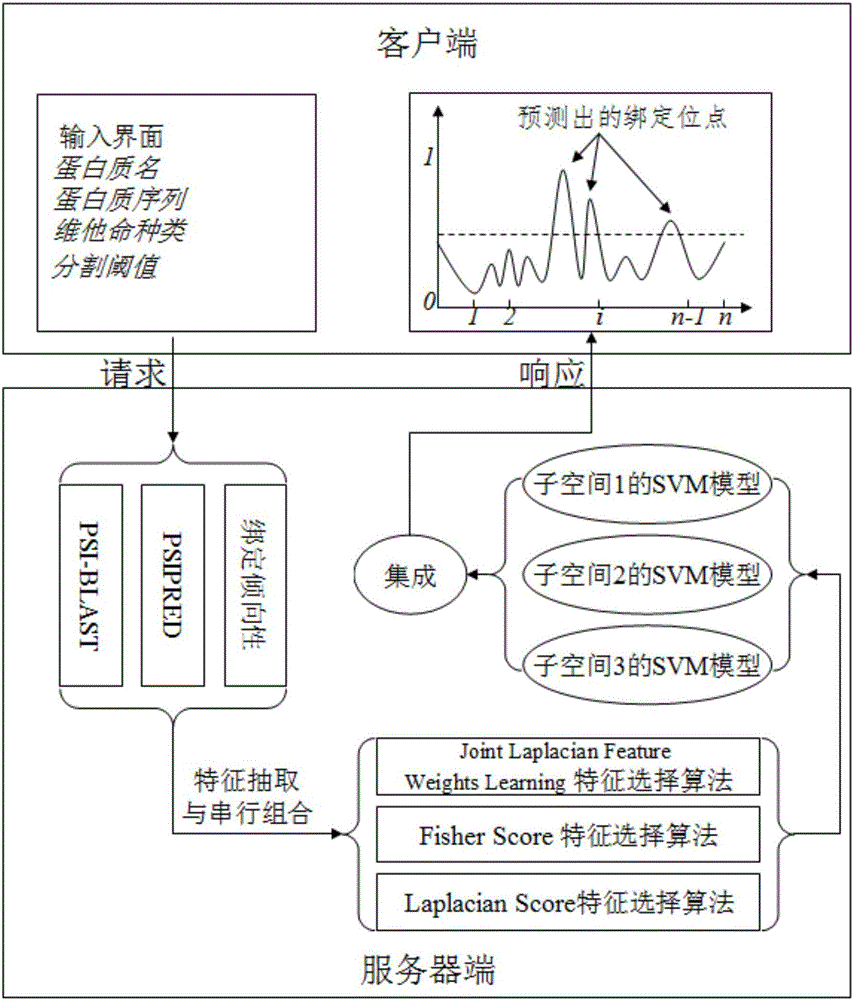
[0086] Such as figure 1 As shown, according to a preferred embodiment of the present invention, the protein-vitamin binding site prediction method based on subspace fusion, first, use PSI-BLAST and PSIPRED to obtain the PSSM matrix (ie evolutionary information matrix) and secondary structure probability matrix, and the binding propensity matrix of the protein generated from the protein-vitamin binding site propensity table; secondly, from the PSSM matrix, the secondary structure probability matrix, and the protein-vitamin binding site The point propensity table was used to construct the feature vector of each amino acid residue; then, the three feature selection algorithms of Joint Laplacian FeatureWeights Learning (Algorithm 1), Fisher Score (Algorithm 2) and Laplacian Score (Algorithm 3) were use...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com