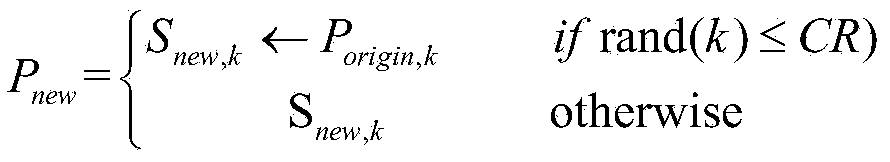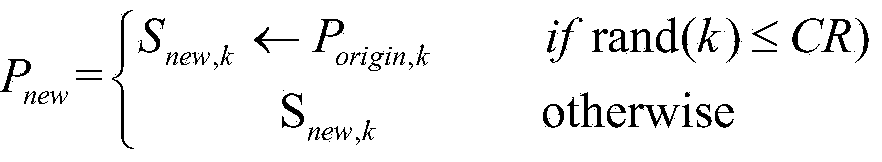Protein conformational space optimization method based on fragment assembly
An optimization method, protein technology, applied in the field of bioinformatics, computer application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033] The present invention will be further described below in conjunction with the accompanying drawings.
[0034] refer to figure 1 , a dynamic step size search method in conformation space, comprising the following steps:
[0035] 1) Initialize the population: Randomly select fragments from the protein fragment library to generate a population P of popSize individuals int , and set the algorithm parameters: population size popSize, protein sequence length Length (that is, the dimension of the optimization problem), the number of iterations T of the algorithm, the crossover factor CR of the algorithm, and the length L of the protein fragment.
[0036] 2) Calculate the function value of each population according to the scoring function f, and sort them, where P max The function value of is optimal.
[0037] 3) When the set termination condition is not reached, perform the following operations
[0038] 3.1) For population P int Each individual P in i Do the following: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com