Nucleic acid aptamer for specific binding with human epidermal growth factor receptor type III variant and application of nucleic acid aptamer
A technology of epidermal growth factor and nucleic acid aptamer, which is applied in the field of biomedicine, can solve the problem of no observed bone marrow suppression, and achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
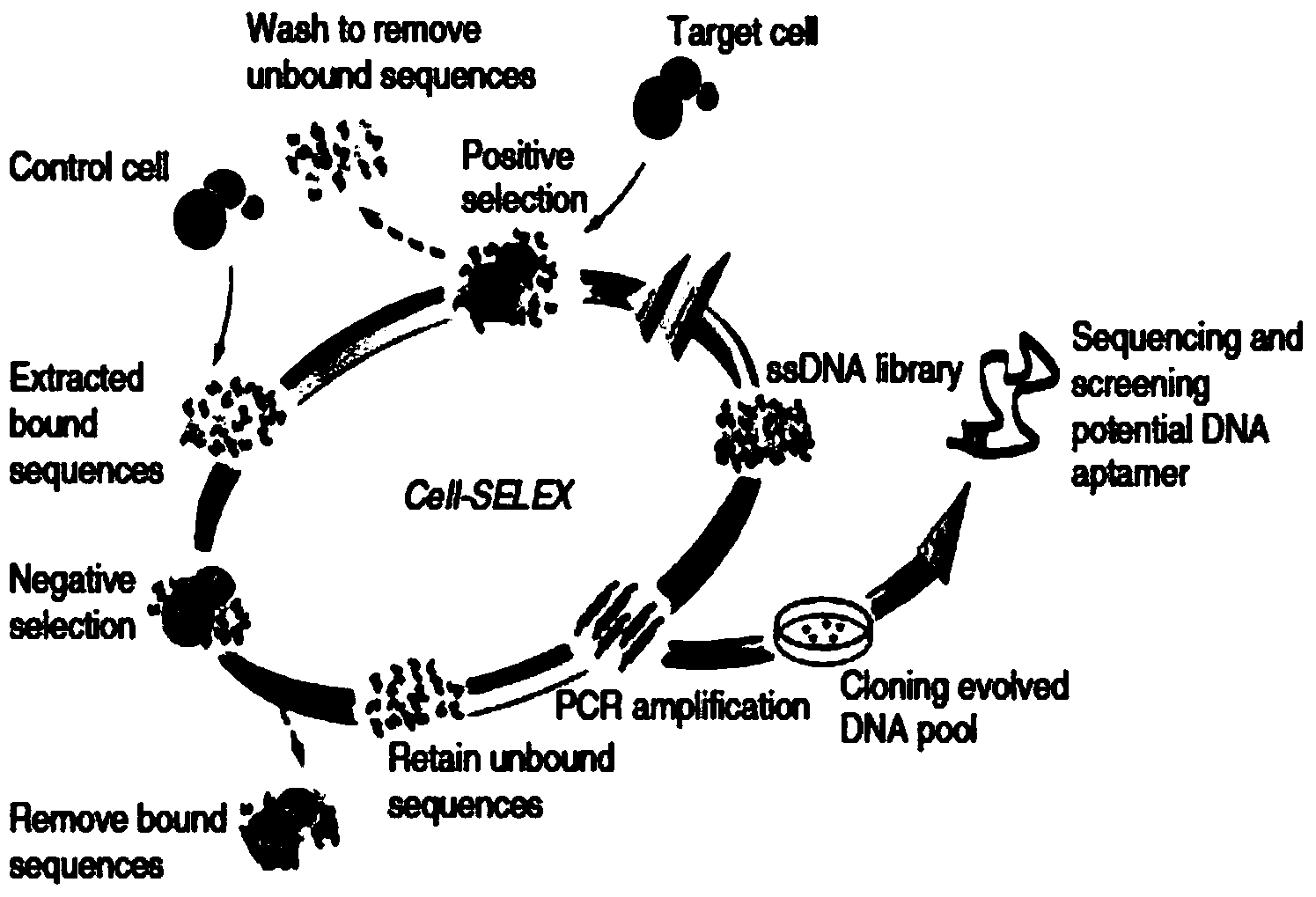
[0056] Experiment 1. Prepare a nucleic acid aptamer that specifically binds to a glioma cell line (U87Δ) overexpressing human epidermal growth factor receptor type III mutant (EGFRvIII). The screening process is as follows: figure 1 As shown, the results of 11 rounds of sequencing are as follows image 3 shown.
[0057] 1. Design and synthesize initial ssDNA library and primers for PCR amplification
[0058] Design and synthesize an initial ssDNA GN library with a length of 76 bases: 5'-ATCCAGAGTGACGCAGCA(N40)TGGACACGGTGGCTTAGT-3'. The two ends of the initial ssDNA library are fixed sequences, and the middle is 40 random sequences. N stands for A, T, C , G four random bases.
[0059] Design and synthesize primers for amplifying double-stranded dsDNA:
[0060] 5'-FITC-Pn1:5'-ATCCAGAGTGACGCAGCA-3';
[0061] 5'-Biotin-Pn2: 5'-ACTAAGCCACCGTGTCCA-3'
[0062] The above-mentioned initial ssDNA library and primers were synthesized by biological companies.
[0063] Using the PCR ...
Embodiment 2
[0099] Experiment 1. The role of aptamers in inhibiting tumor growth in nude mouse tumor-bearing animal models
[0100] Specific method: U87Δ cells were treated with DMEM containing 10% FBS in 5% CO 2 , Cultured at 37°C, and nude mice were raised in the SPF experimental barrier environment for 4 to 6 weeks. A total of 28 nude mice were divided into blank group and in vivo jet-PEI TM Transfection reagent group, original library GN group and aptamer U2 treatment group, 4 groups, 7 animals in each group; U87Δ cells were resuspended in modified RPMI-1640 culture medium, and 1.5×10 6 ~2.0×10 6 Use a 1ml syringe to subcutaneously inject 100 μl into the right hip of male nude mice (BALB / c) at 4 to 6 weeks old; observe the tumor formation after 2 weeks, and measure the tumor volume with a vernier caliper; use in vivo jet-PEI TM The reagents were transfected with the aptamer U2 and the original library GN (initial ssDNA library) for intratumoral injection (once every other day, 67p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com