Identification method for amblyopyrum muticum chromosome in wheat genome
A technology of goat grass and identification method, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., which can solve the problems of cumbersome operation steps, long and short length of blocked DNA, etc., and broaden the scope of application Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
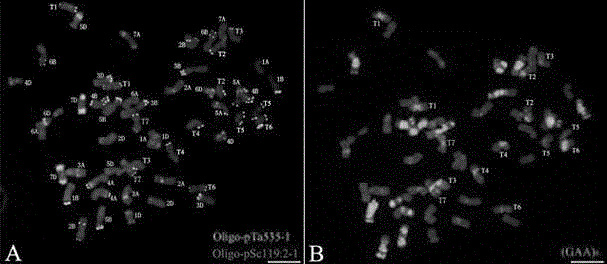
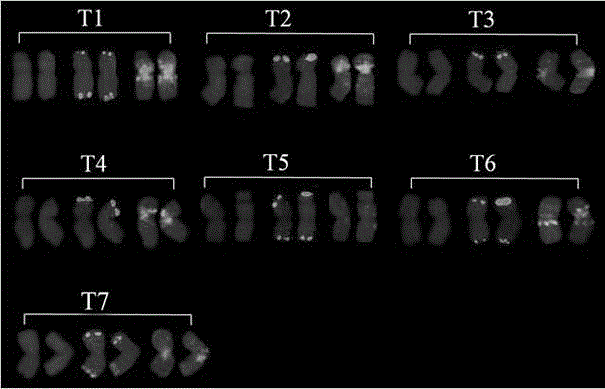
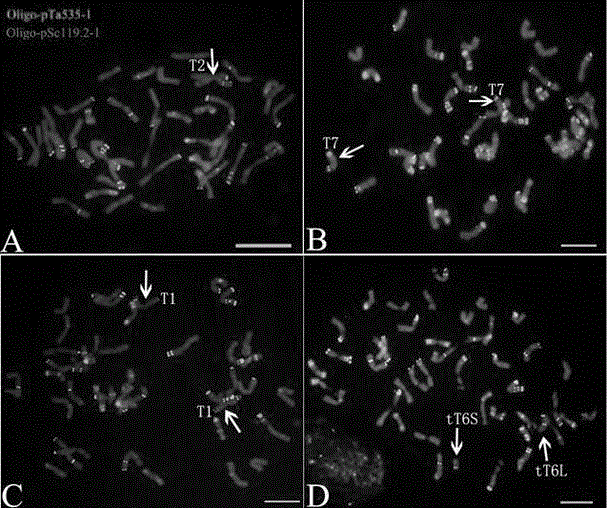
[0043] Hybrid F 1 ; Hybrid F 1 After doubling, the innovative germplasm of Chinese spring-goat grass (No. XX001) was obtained. The material numbered XX002-XX031 is the hybrid backcross of XX001 and CS (BC 2 f 5 ) offspring. Wheat China Spring (CS) was provided by Professor Yang Zujun, School of Life Science and Technology, University of Electronic Science and Technology of China. Goatgrass (TA2193) was provided by Dr. Raupp J, Center for Wheat Genomics and Genetic Resources, Kansas State University, USA. Probe Oligo-pSc119.2-1, probe Oligo-pTa535-1 and (GAA)8 were synthesized by Chengdu Ruixin Biological Company. The 3 probe sequences are as follows:
[0044] Probe Oligo-pSc119.2-1 sequence:
[0045] CCGTTTTGTGGACTATTACTCACCGCTTTGGGGTCCCATAGCTAT, as shown in SEQ ID NO.1;
[0046] Probe Oligo-pTa535-1 sequence:
[0047] AAAAACTTGACGCACGTCACGTACAAATTGGACAAACTCTTTCGGAGTATCAGGGTTTC, as shown in SEQ ID NO.2;
[0048] Probe (GAA) 8 sequence: GAAGAAGAAGAAGAAGAAGAAGAA, as show...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com