Rca reporter probes and their use in detecting nucleic acid molecules
A probe and molecule technology, applied in the field of detection of analytes such as target nucleic acid molecules and nucleic acid molecules, can solve the problems of increasing time and resources, difficulty in automation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0314] Example 1 - Two-Part Hairpin RCA Probes
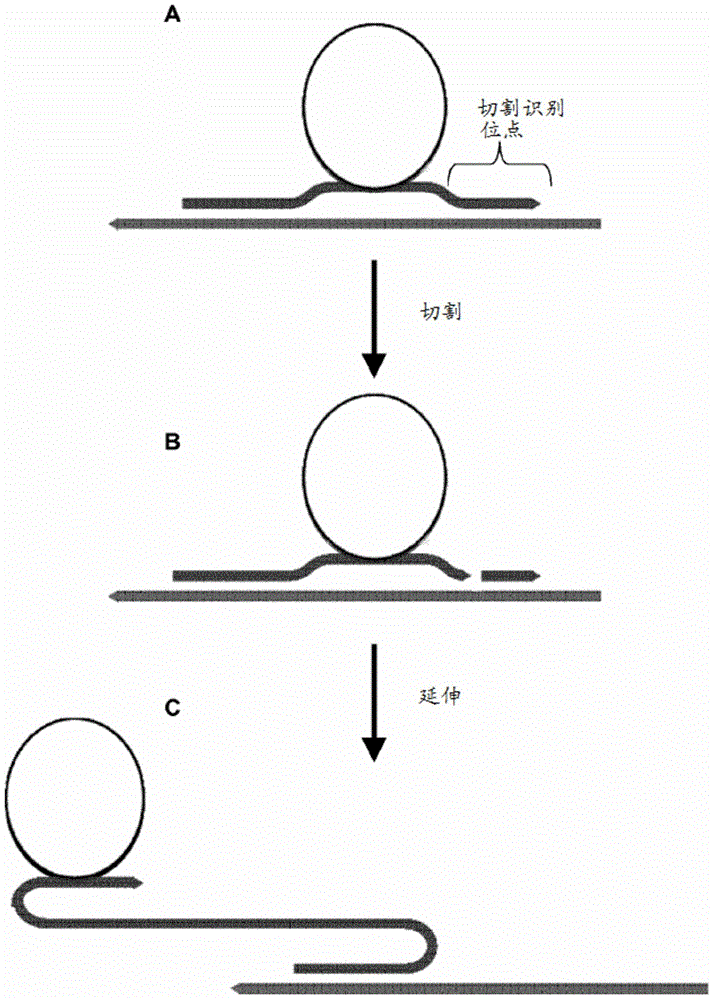
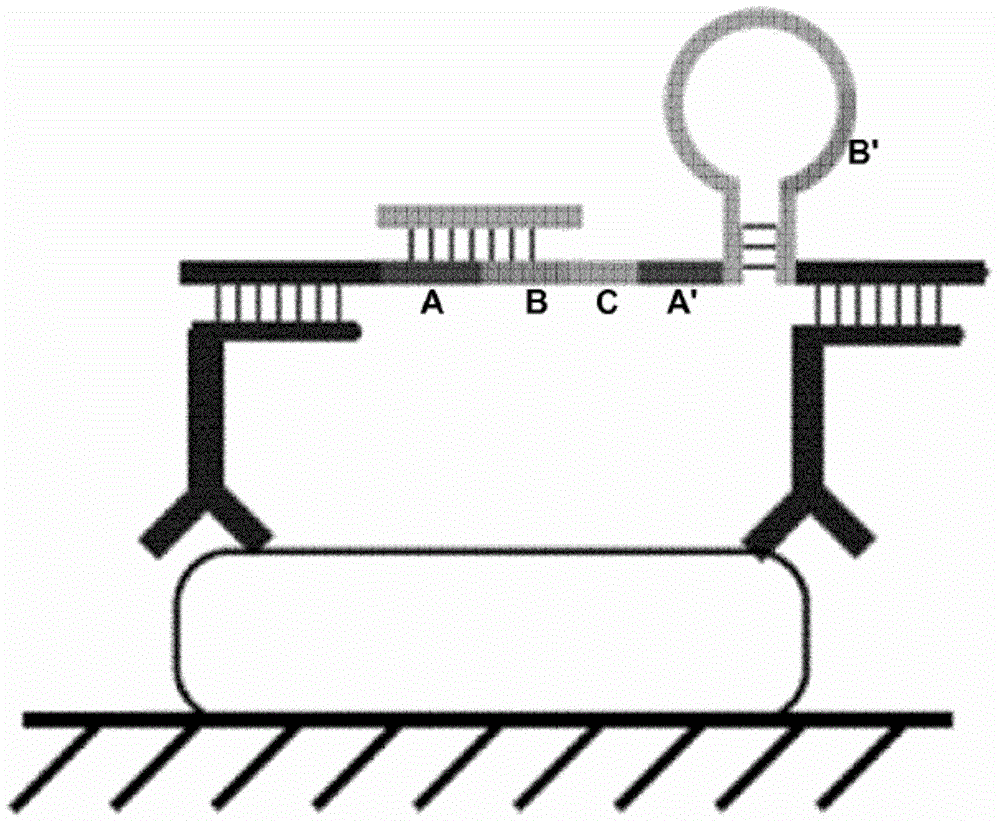
[0315] As noted above, the probes of the invention can be used to detect any analyte wherein an intermediate binding partner that can bind to the analyte is coupled to (or comprises) a nucleic acid molecule. A nucleic acid molecule coupled to an intermediate molecule, which serves as a surrogate or marker for the analyte, can be detected using the probe. The protocol described hereinafter uses a proximity probe as an intermediate binding molecule to a target analyte, wherein the target complementary domain of the RCA probe binds to the nucleic acid domain of the proximity probe (see e.g. figure 2). RCA probes were compared to padlock probes using gap oligonucleotides that are also capable of binding to the nucleic acid domains of the proximity probes to generate RCA templates. The RCA reaction of the padlock probe RCA template is initiated by one of the proximity probe nucleic acid domains.
[0316] 50 μl of biotinylated m...
Embodiment 2- 3
[0323] Example 2 - Three-Part Hairpin RCA Probe
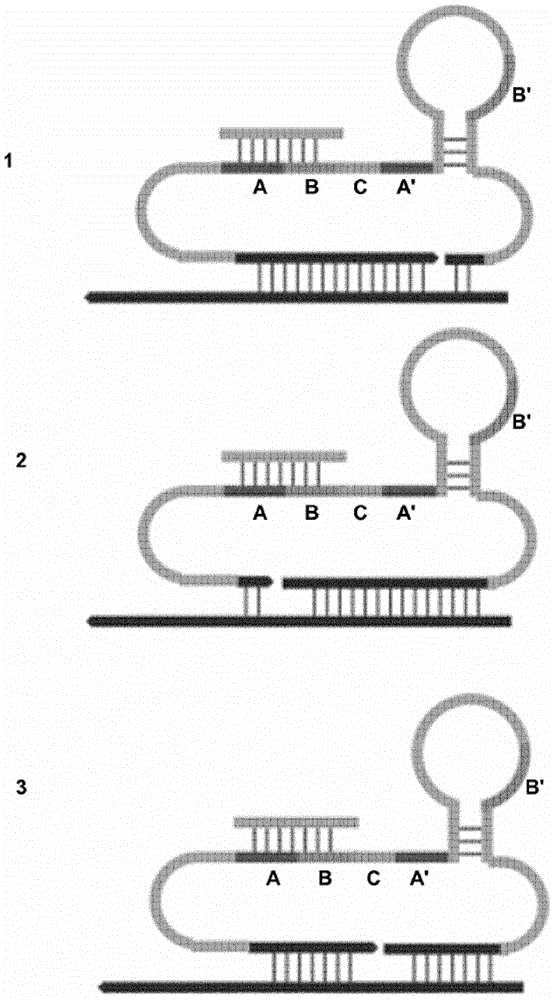
[0324] The utility of "triple-part" RCA probes was demonstrated using nucleic acid analytes, ie, the target analyte is a nucleic acid molecule. Two variants of "triple-part" probes were tested, namely single-stranded probes containing 3 hairpin structures and partially double-stranded probes containing hairpin structures and two cleaved strands.
[0325] 50 μl of 10 pM synthetic biotinylated DNA template was incubated on streptavidin-coated slides (SurModics) in 1×PBS at 37° C. for 60 minutes, with 1×PBS without DNA incorporated as a negative control. After incubation, block with blocking buffer (0.1% BSA (Sigma), 100nM goat IgG (Sigma), 1mM biotin (Sigma), 10ng / μl salmon sperm DNA (Sigma), 5mM EDTA, 1x PBS and 0.05% Tween 20) Slides were blocked at 37°C for 60 minutes.
[0326] Streptavidin-coated slides were compartmentalized with secure-Seal 8 (Grace Bio-labs) prior to the experiment. A wash step (with two replicates of...
Embodiment 3
[0331] Continuous two-part probe and cut strand two-part probe in the detection of embodiment 3-nucleic acid target
[0332] The utility of "two-part" RCA probes is demonstrated using nucleic acid analytes, ie the target analyte is a nucleic acid molecule. Target nucleic acids were immobilized on glass slides using two different techniques. Two variants of "two-part" probes were tested, namely a continuous probe comprising two hairpin structures and a partially double-stranded probe comprising both a hairpin structure and a cleaved strand.
[0333] Immobilization of synthetic DNA templates
[0334] 10 [mu]l of synthetic biotinylated DNA template in 1x PBS was exposed to streptavidin-coated slides (SurModics) for 10 min at 55[deg.]C. Alternatively, hybridize the DNA template to the slide in 50 μl 1x PBS for 60 minutes at 37°C, using 1x PBS as a negative control. After incubation, at 37 ° C with blocking buffer (0.1% BSA (Sigma), 100nM goat IgG (Sigma), 1mM biotin (Sigma), ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com