Method for detecting EGFRvIII in tumor tissues
A technology for tumor tissue and tissue samples, applied in the field of molecular biology, can solve the problems of complex operation, cost limitation, and long time consumption, and achieve the effect of short time consumption, high accuracy, and guaranteed accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0024] The following will clearly and completely describe the technical solutions of the embodiments of the present invention in conjunction with the accompanying drawings. Apparently, the described embodiments are only some of the embodiments of the present invention, not all of them.
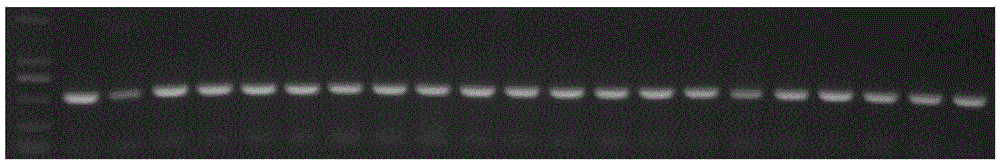
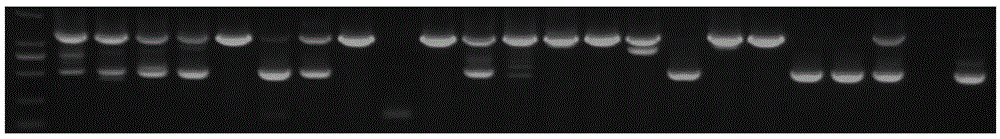
[0025] A method for detecting EGFRvIII in tumor tissue according to an embodiment of the present invention, comprising the following steps:
[0026] 1. EGFRvIII gene sequence analysis and primer design
[0027] Use the written sequence analysis program to scan the gene sequence before the mutation region (71.6% GC content, high repetition rate) base by base to obtain a low-repetition region of 56-70 bases, and use the sequence analysis software to design a specific reaction. The forward primers were matched with the forward primers F56 (SEQ ID NO: 1) and F66 (SEQ ID NO: 3) before the mutation region, and the specificity of each paired primer in the human genome was detected by using the NCBI p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com