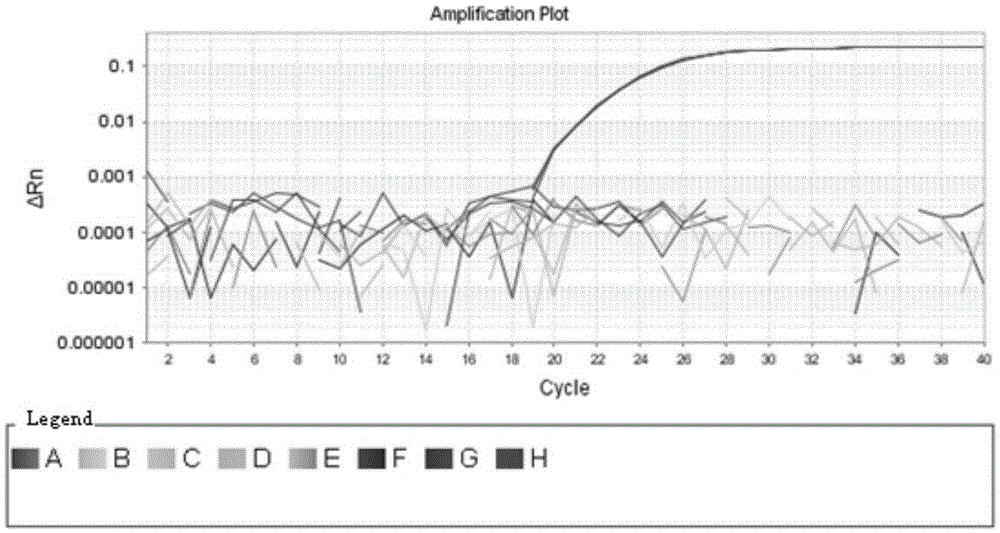
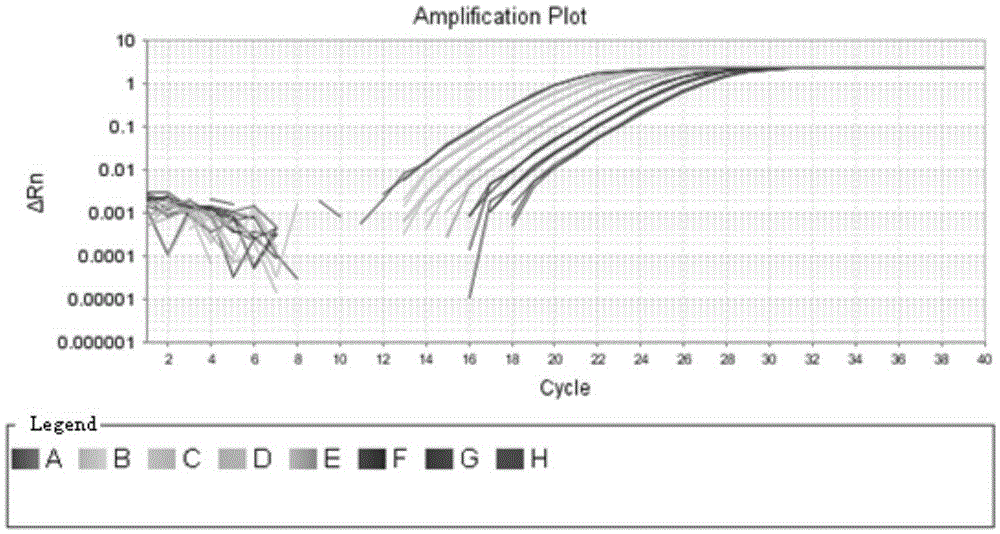
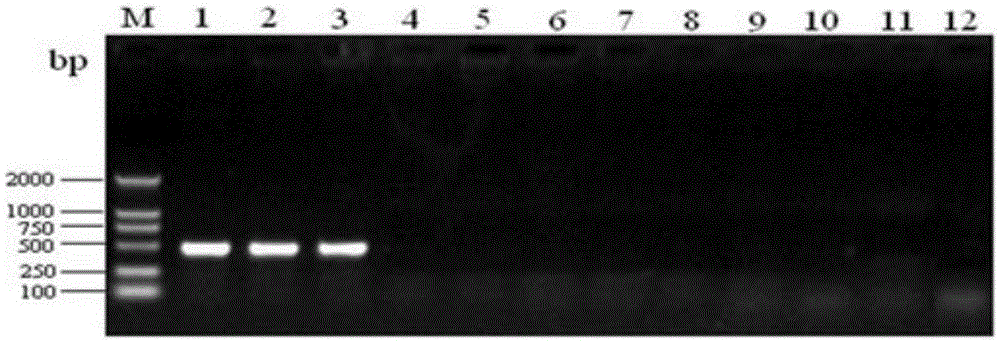
Rapid qualitative and quantitative detection kit for enterococcus faecium in feed as well as detection method and application
A technology for quantitative detection of Enterococcus faecium, applied in the direction of microbial determination/testing, biochemical equipment and methods, etc., can solve the problem that qualitative and quantitative detection methods are not scientific and fast enough, limit the development of probiotic preparations, and consume a lot of time and effort Resources and other issues, to achieve the effect of low detection standards, high sensitivity, and good accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Selection of target detection genes for Enterococcus faecium:
[0027] The present invention is effective for other probiotics such as Lactobacillus acidophilus, Lactobacillus bulgaricus, Lactobacillus plantarum, Lactobacillus bulgaricus, Lactobacillus casei, Enterococcus faecalis, Bifidobacterium, etc. A large number of comparisons and analyzes were carried out to select the target gene for qualitative and quantitative detection. The target gene is a conserved housekeeping gene in the genome of Enterococcus faecium, and it is a single-copy gene. The conserved sequence of the gene was designed Specific primers are used for qualitative and quantitative detection of Enterococcus faecium in feed, and the obtained data can truly reflect the quantity of Enterococcus faecium added in feed. The primers designed for the target gene are: 5'-GCGTCCAAGTTATTTTCCCATTTAC-3', 5'-CGATTCCTGACATCTTTCAACACA-3', which can be used for qualitative and quantitative detection of Enterococcus f...
Embodiment 2
[0029] Rapid qualitative and quantitative detection kit for Enterococcus faecium added in feed, including primers: 5'-GCGTCCAAGTTATTTTCCCATTTAC-3', 5'-CGATTCCTGACATCTTTCAACACA-3.
Embodiment 3
[0031] The detection method for the rapid qualitative and quantitative detection kit of Enterococcus faecium added in feed, including the method for pretreatment of feed samples, the method includes:
[0032] Mix 25g of the fully pulverized and homogeneous feed with 225mL of sterilized physiological saline, shake at 4°C for 1-2h at a speed of 100 rpm, and prepare a 10% uniform dilution. Aseptically perform 10-fold incremental dilution on the homogeneous dilution prepared in the previous step, select more than 2 to 3 appropriate dilutions, and extract bacterial genomic DNA or RNA as needed.
[0033] The remaining steps utilize the primers in Example 1 to carry out qualitative and quantitative identification of the extracted DNA or RNA. The detection method for the rapid qualitative and quantitative detection kit of Enterococcus faecium added in the feed specifically includes the following steps:
[0034] A. Preparation of template DNA
[0035] Aseptically put 25g of feed that...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com