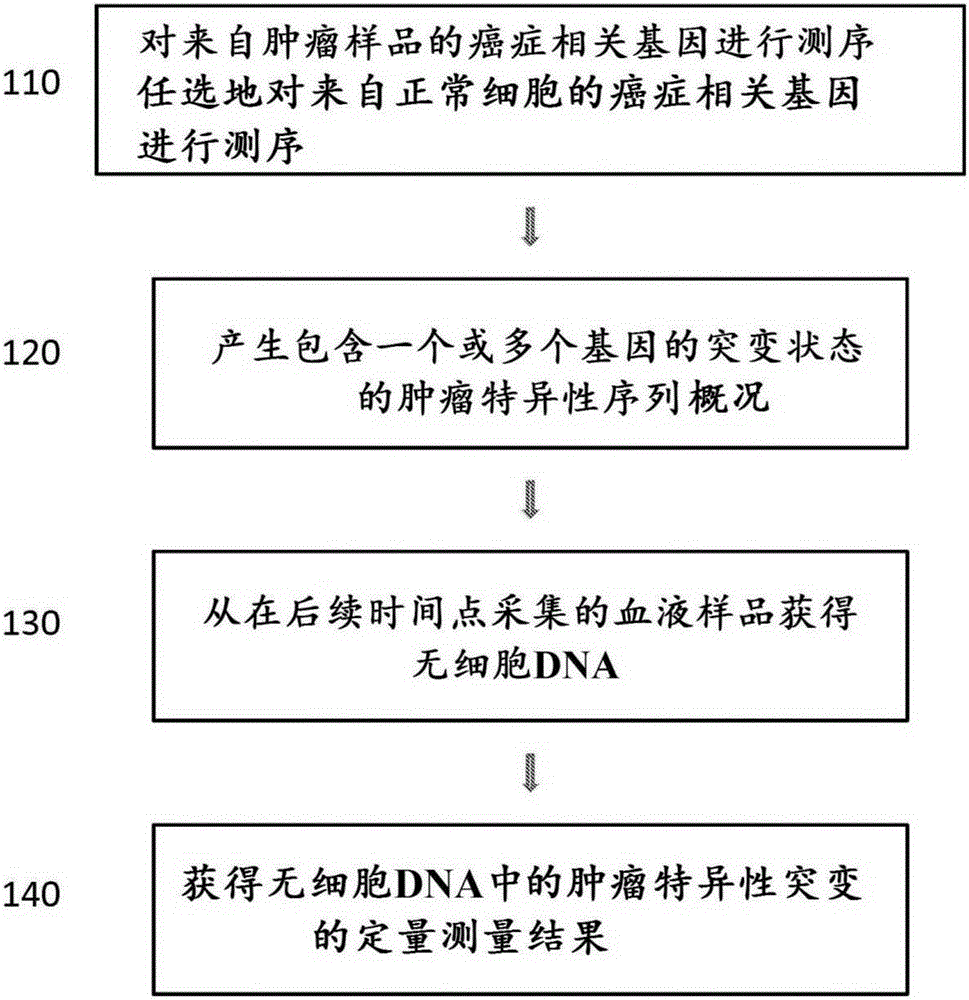
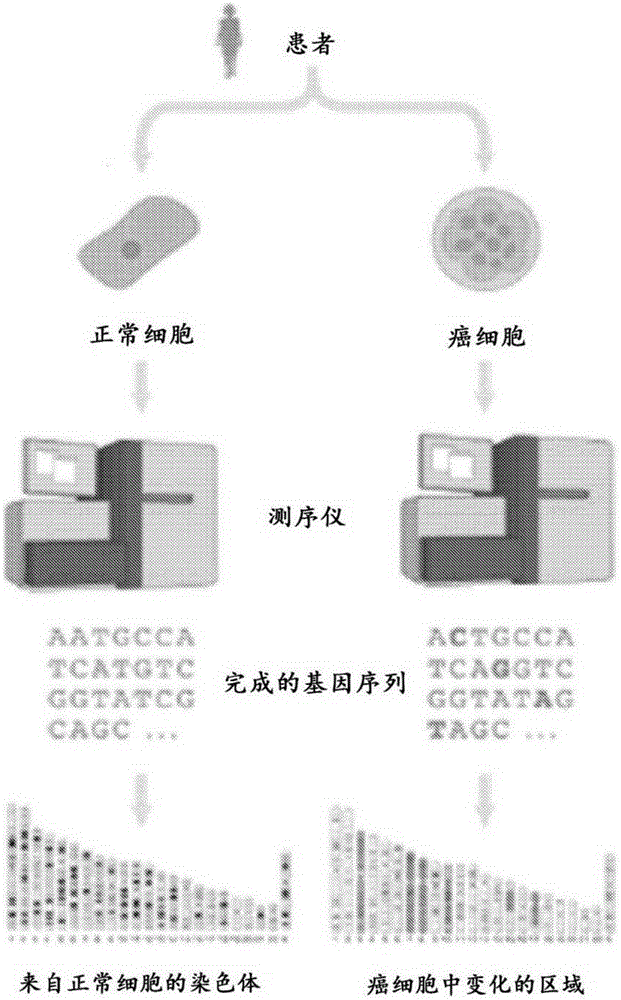
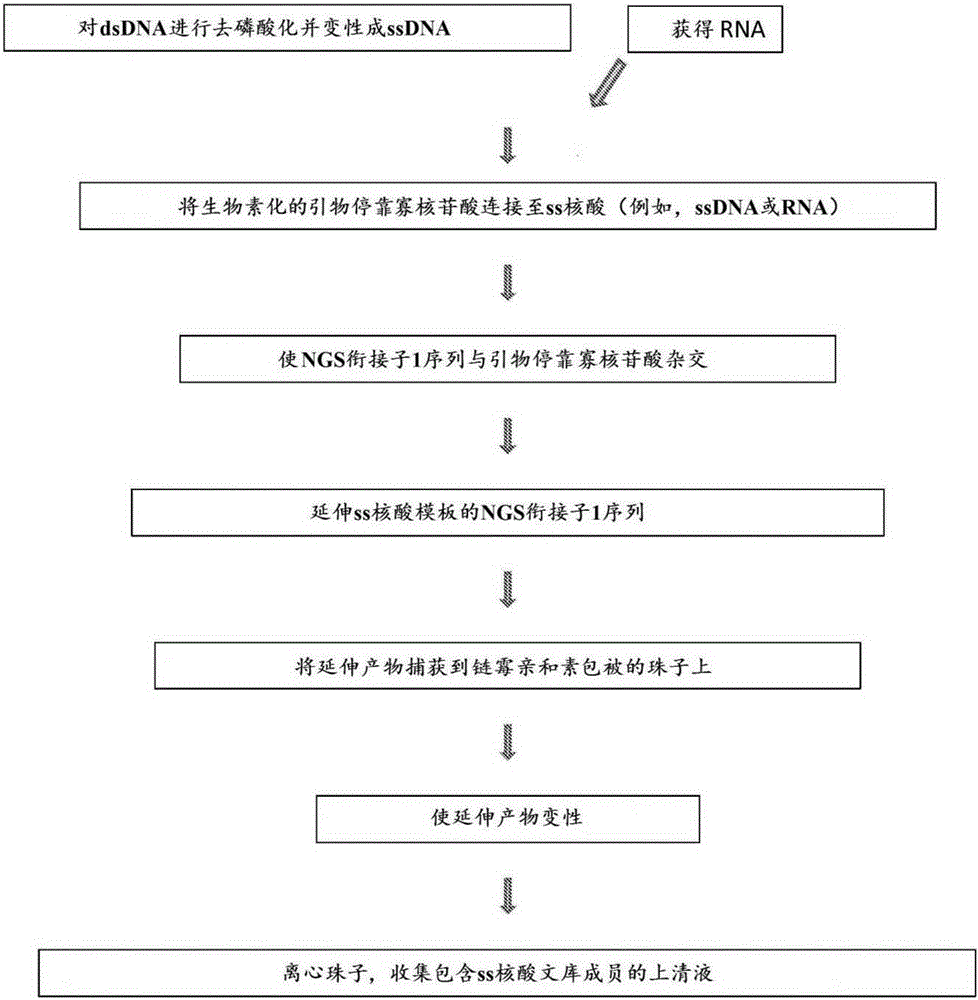
Methods, compositions, and kits for nucleic acid analysis
An oligonucleotide and subgroup technology, applied in combinatorial chemistry, nucleotide library, organic compound library, etc., can solve problems such as poor application of diagnostic sequencing and sequencing errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0410] Preparation of target-enriched libraries
[0411] In another aspect, the present invention provides a method for preparing a target-enriched DNA library. The method can include hybridizing target-selective oligonucleotides to sequencing library members to produce hybridization products. The method may further comprise amplifying the hybridization product in a single round of amplification to generate an extended strand.
[0412] Methods of target enrichment can be as described in US Patent Application Publication No. 20120157322, which is incorporated herein by reference.
[0413] Hybridization and amplification can occur in the reaction mixture. The mixture may contain nucleotides (dNTPs), polymerases, and target-selective oligonucleotides. In some embodiments, the mixture comprises a plurality of target-selective oligonucleotides. The mixture may contain, for example, 1-10, 5-20, 10-50, 40-100, 80-200, 150-500, 300-1000, 800-2000, 1000-5000, 4000-10000, 8000-200...
Embodiment 1
[0536] Figure 24Methods for assessing cancer in a subject are shown. The subject has had a colonoscopy and was found to have a colon tumor. At time point 0, tumor biopsies and blood draws were taken from subjects and used to aid in the diagnosis of colon cancer in subjects. Tumor and normal cells from the first blood draw were sequenced. Sequencing revealed the presence of three mutations in the subject's tumor. The mutation is a point mutation in the APC, KRAS and TP53 genes. The stage of the subject's cancer was determined. The subject underwent the first treatment (surgery) to remove the tumor. During the first treatment, a second blood draw was performed. The subject's tumor is determined to have metastasized. The subject is administered a second therapy (chemotherapy) to control the cancer. Subsequent blood draws were performed to determine the mutational status of these three genes in cell-free DNA from blood.
Embodiment 2
[0537] Example 2: Validation assay for tumor-specific mutations in subjects with colon cancer
[0538] The NCI-H1573 (CRL-5877) cell line with the KRASG12A mutation (mu) was obtained as a frozen stock from the American Type Culture Collection (ATCC). Genomic DNA (gDNA) was prepared from cell line material using a commercially available kit (DNeasyBlood & Tissue Kit, QIAGEN) according to the manufacturer's suggested protocol. Estimates of DNA concentration were obtained spectrophotometrically by measuring OD260 (NanoDrop1000, ThermoFisher Scientific Inc.).
[0539] Genomic DNA from the NA18507 cell line was used as a surrogate for wild-type DNA (wt) and was obtained as a purified stock (Coriell). Combine two microliters of the mixture containing wt (30 ng) and mu (6 ng) DNA into a 20 μl ddPCR reaction mix from a 2x ddPCR supermix for probes at a final concentration of 0.2 uM of each Forward primer (wt: 5'-AGATTACGCGGCAATAAGGCTCGGTTGGCATTGGATACTACTTGCCTACGCCACC-3' (SEQ ID NO: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com