Super-long genome-based variation detection algorithm and detection system
A mutation detection and genome technology, applied in the direction of microbial determination/inspection, calculation, biochemical equipment and methods, etc., can solve the problem of inability to accurately detect the structural variation of large and ultra-long genomes, and achieve the effect of improving sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0067] The principles and features of the present invention are described below in conjunction with the accompanying drawings, and the examples given are only used to explain the present invention, and are not intended to limit the scope of the present invention.
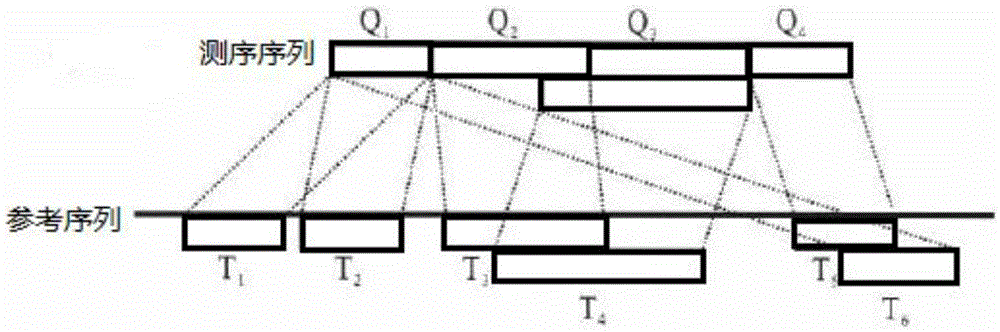
[0068] Such as figure 1 As shown, a variation detection algorithm based on ultra-long genomes includes the following steps:
[0069] S1. Using a partial sequence alignment algorithm to detect all matches between the sequencing fragments and the reference sequence, and obtain partial matching events, each partial matching event includes the sequencing fragments and the reference fragments on the reference sequence;
[0070] S2. Sorting the sequenced fragments in all partial matching events according to the positions compared to the reference sequence, and grouping the partial matching events whose positions on the reference sequence overlap or are sequentially connected by the sequenced fragments;
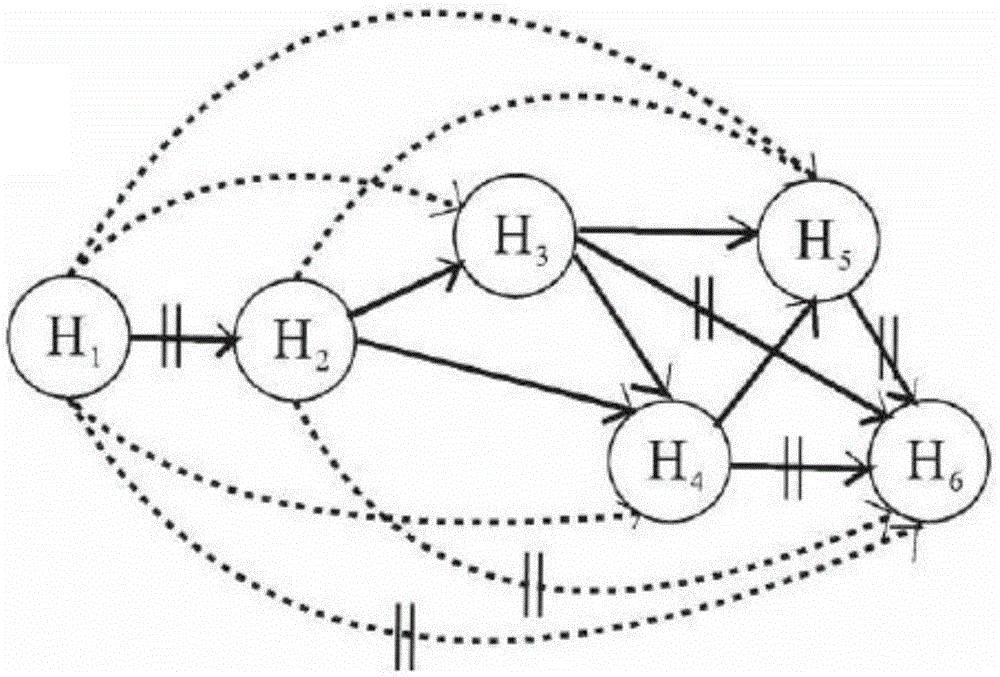
[0071] S3. Score...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com