Reverse transcription fluorogenic quantitative PCR primer for rapidly identifying high virulent Newcastle disease virus (NDV) strain and identification method
A fluorescence quantitative, virulent strain technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as high cost and poor sensitivity, and achieve simple and specific detection methods. , the effect of high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1: Optimization of RT-qPCR reaction conditions
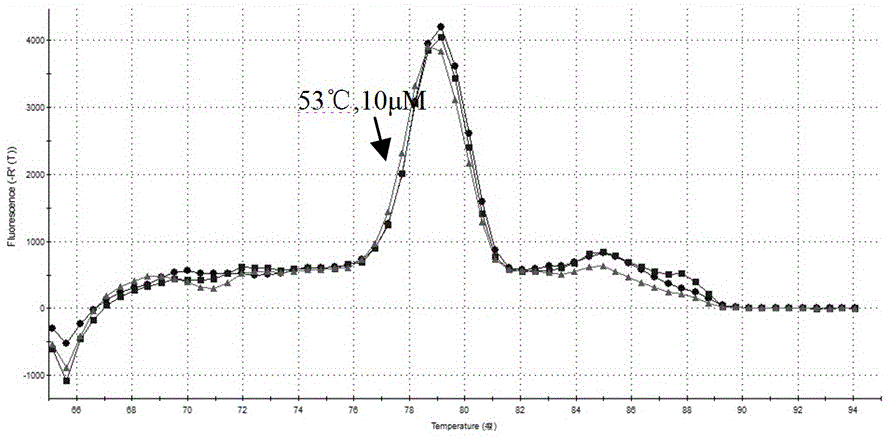
[0034] The determination of the reaction conditions is mainly to optimize the primer concentration and annealing temperature. The reaction system is 20 μL, including 10 μL of 2×OneStepSYBRRT-PCRBuffer; 0.4 μL of ExTaqHS; 0.4 μL of PrimescriptrtenzymemixⅡ; 1 μL of NDVFrealtime-1 and NDVFrealtime-2 primers (5-100 μM); RnaseFreedH 2 O5.2μL; TotalRNA2μL. The reaction conditions were: reverse transcription at 42°C for 5 minutes; pre-denaturation at 95°C for 3 minutes; 40 cycles of amplification at 94°C for 10s, 51-57°C for 10s, and 72°C for 20s, and fluorescence signals were collected for each cycle. The primer concentration was 10 μM, 20 μM, 30 μM, 40 μM, and 50 μM in 5 concentration gradients, and the annealing temperature was 51 °C, 53 °C, 54 °C, 55 °C, and 57 °C at 5 reaction temperatures, and each reaction condition was repeated three times. . The results were judged according to the amplification curve and ...
Embodiment 2
[0035] Embodiment 2: establish standard curve
[0036] According to the determined optimal reaction conditions, with 10 8 、10 7 、10 6 、10 5 、10 4 、10 3 Copy / μL RNA standards with 6 concentration gradients were used as templates for the establishment of RT-qPCR standard curves. The reaction system of each sample is the same, 20 μL, including 10 μL of 2×OneStepSYBRRT-PCRBuffer; 0.4 μL of ExTaqHS; 0.4 μL of PrimescriptrtenzymemixⅡ; 2 O5.2μL; TotalRNA2μL.
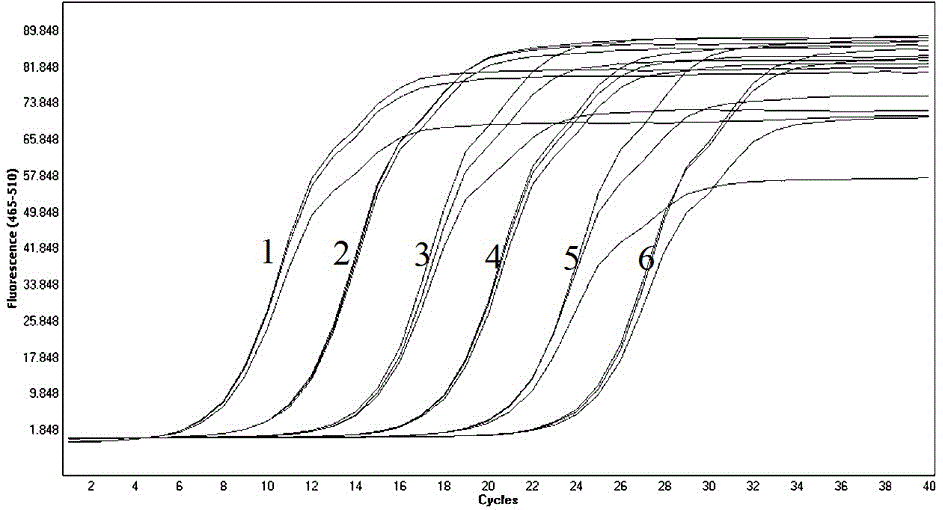
[0037] The reaction conditions were determined as follows: reverse transcription at 42°C for 5 minutes; pre-denaturation at 95°C for 3 minutes; 40 cycles of amplification at 94°C for 10s, 53°C for 10s, and 72°C for 20s. repeated to obtain the respective kinetic curves, such as image 3 . Then obtain the standard curve, the linear equation of the standard curve is Y=-3.307X+2.657, such as Figure 4 . The results showed that the repeatability of each dilution was good, and the established standard curve was qualified. ...
Embodiment 3
[0038] Embodiment 3: sensitivity test
[0039] respectively by 10 6 、10 5 、10 4 、10 3 、10 2 、10 1 、10 0 The copy / μL standard product is used as a template, and other conditions are the same as in Example 2, and RT-qPCR amplification is performed, and the detection limit value of the copy number detection method of the lowest dilution that can be effectively amplified. The results showed that the minimum concentration of the template that could be detected by fluorescent quantitative PCR was 10 0 copy / μL( Figure 5 ), which indicated that the detection limit of RT-qPCR was 1 copy / μL.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com