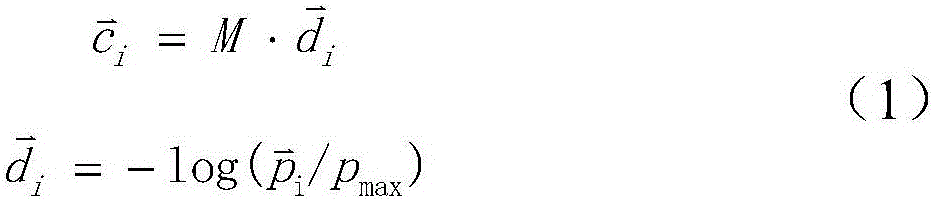Pathological image cell nucleus quick location method
A pathological image and positioning method technology, applied in the field of digital image processing of cell nucleus center positioning in digital pathological images, can solve the problem that the processing speed is difficult to meet the computer-aided analysis of digital pathological full slices, and achieve easy implementation, fast execution speed, and flow clear effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0016] In order to better understand the technical solution of the present invention, the present invention will be described in detail below in conjunction with the accompanying drawings and specific embodiments.
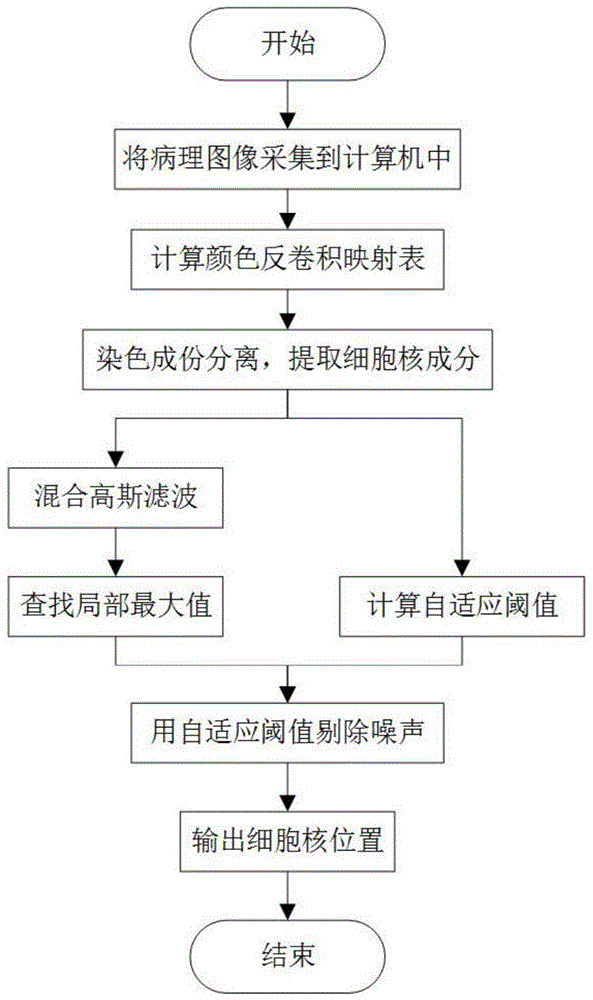
[0017] The present invention is a method for rapidly locating cell nuclei in pathological images, and the method mainly includes the following steps:
[0018] 1. Use a slice scanner to scan pathological slices for tissue biopsy into an electronic computer, and store them as a digital image matrix in the form of RGB three-channel.
[0019] 2. Use the color deconvolution matrix to calculate the color deconvolution mapping table.
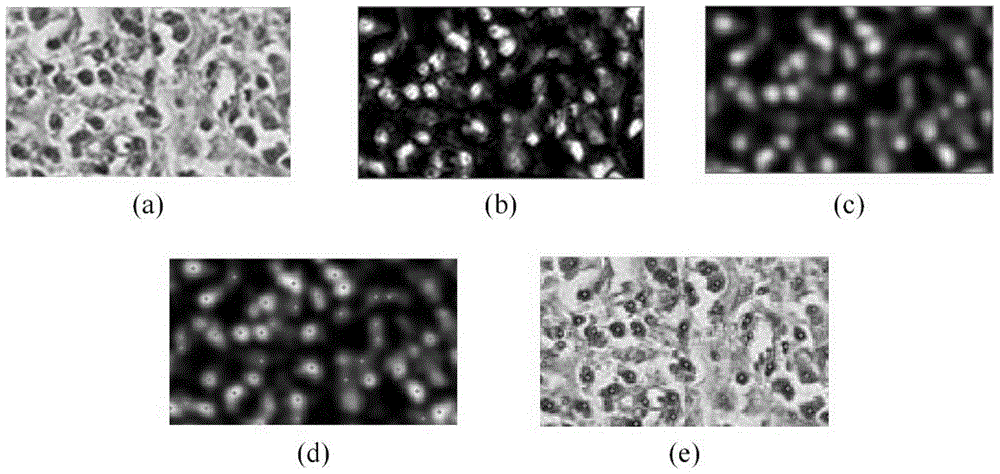
[0020] 3. Using the color deconvolution mapping table in step 2 to extract the nuclei components in the digital pathological slice, and obtain the nuclei component images.
[0021] 4. Perform mixed Gaussian filtering on the nucleus component image obtained in step 3 to obtain a filtered image.
[0022] 5. Find the local maximum in the fil...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com