Primer combination for detecting mutation of PIK3CA gene in trace tissue and application of primer combination
A primer combination and primer set technology, applied in recombinant DNA technology, microbial determination/inspection, biochemical equipment and methods, etc., can solve the problem of insufficient tissue sample acquisition, complicated operation, and low specificity of target sequence amplification, etc. problem, to achieve the effect of good practical application value, high amplification efficiency and low price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
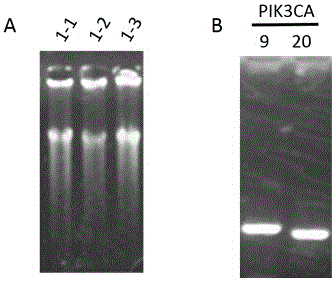
[0031] Example 1: In the DNA template of 1000pgPIK3CA Gene Mutation Detection
[0032] 1. Experimental materials
[0033] phi29 DNA polymerase, 10×reaction buffer, dNTP (10nM), 100×BSA, 1000pg DNA template, deionized water, primer1-prime18 (Primer Mix), 2×Pfu Mix, primer19-primer22, PCR machine, super Thin product purification kit (Tiangen Biochemical Technology (Beijing) Co., Ltd. DP203), 1% agarose gel, electrophoresis apparatus.
[0034] 2. Experimental procedures and results
[0035] 2.1 Pre-amplification
[0036] (1) Primer dilution
[0037] Primer1-prime18 was diluted and mixed to form a Primer Mix (Table 1), wherein the final concentration of primer1-primer16 primers was 0.1 μM, and the final concentration of primer17-primer18 primers was 2 μM. Configure the following system:
[0038] ;
[0039] (2) After mixing the above reagents, pre-denature at 98°C for 10 minutes, and then quickly put them on ice for 20 minutes;
[0040] (3) Then add the following system: ...
Embodiment 2
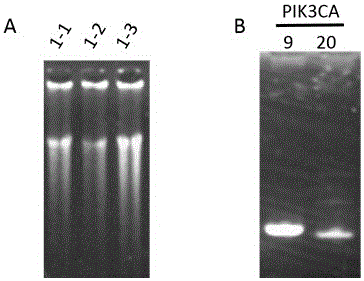
[0057] Embodiment 2: In the DNA template of 500pg PIK3CA Gene Mutation Detection
[0058] 1. Experimental materials
[0059] phi29 DNA polymerase, 10×reaction buffer, dNTP (10nM), 100×BSA, 1000pg DNA template, deionized water, primer1-prime18 (Primer Mix), 2×Pfu Mix, primer19-primer22, PCR machine, super Thin product purification kit (Tiangen Biochemical Technology (Beijing) Co., Ltd. DP203), 1% agarose gel, electrophoresis apparatus.
[0060] 2. Experimental steps
[0061] 2.1 Pre-amplification
[0062] (1) Primer dilution. Primer1-prime18 was diluted and mixed to form a Primer Mix (Table 1), wherein the final concentration of primer1-primer16 primers was 0.1 μM, and the final concentration of primer17-primer18 primers was 2 μM. Configure the following system:
[0063]
[0064] (2) After mixing the above reagents, pre-denature at 98°C for 10 minutes, and then quickly put it on ice for 20 minutes.
[0065] (3) Then add the following system:
[0066]
[0067] Afte...
Embodiment 3
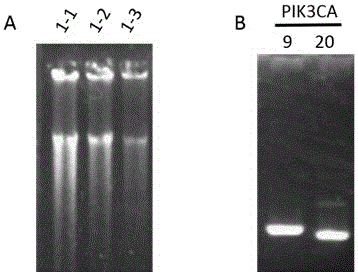
[0081] Example 3: In the DNA template of 100pg PIK3CA Gene Mutation Detection
[0082] 1. Experimental materials
[0083]phi29 DNA polymerase, 10×reaction buffer, dNTP (10nM), 100×BSA, 1000pg DNA template, deionized water, primer1-prime18 (Primer Mix), 2×Pfu Mix, primer19-primer22, PCR machine, super Thin product purification kit (Tiangen Biochemical Technology (Beijing) Co., Ltd. DP203), 1% agarose gel, electrophoresis apparatus.
[0084] 2. Experimental steps
[0085] 2.1 Pre-amplification
[0086] (1) Primer dilution. Primer1-prime18 was diluted and mixed to form a Primer Mix (Table 1), wherein the final concentration of primer1-primer16 primers was 0.1 μM, and the final concentration of primer17-primer18 primers was 2 μM.
[0087] Configure the following system:
[0088]
[0089] (2) After mixing the above reagents, pre-denature at 98°C for 10 minutes, and then quickly put it on ice for 20 minutes.
[0090] (3) Then add the following system:
[0091]
[0092]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com