Method for gene editing and expression regulation using split cas system
一种基因编辑、编码基因的技术,应用在化学仪器和方法、生物化学设备和方法、基因工程等方向,能够解决Cas9基因编辑效率低等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0111] Example 1. Obtaining and functional verification of split Cas9 protein group and recombinant vector group
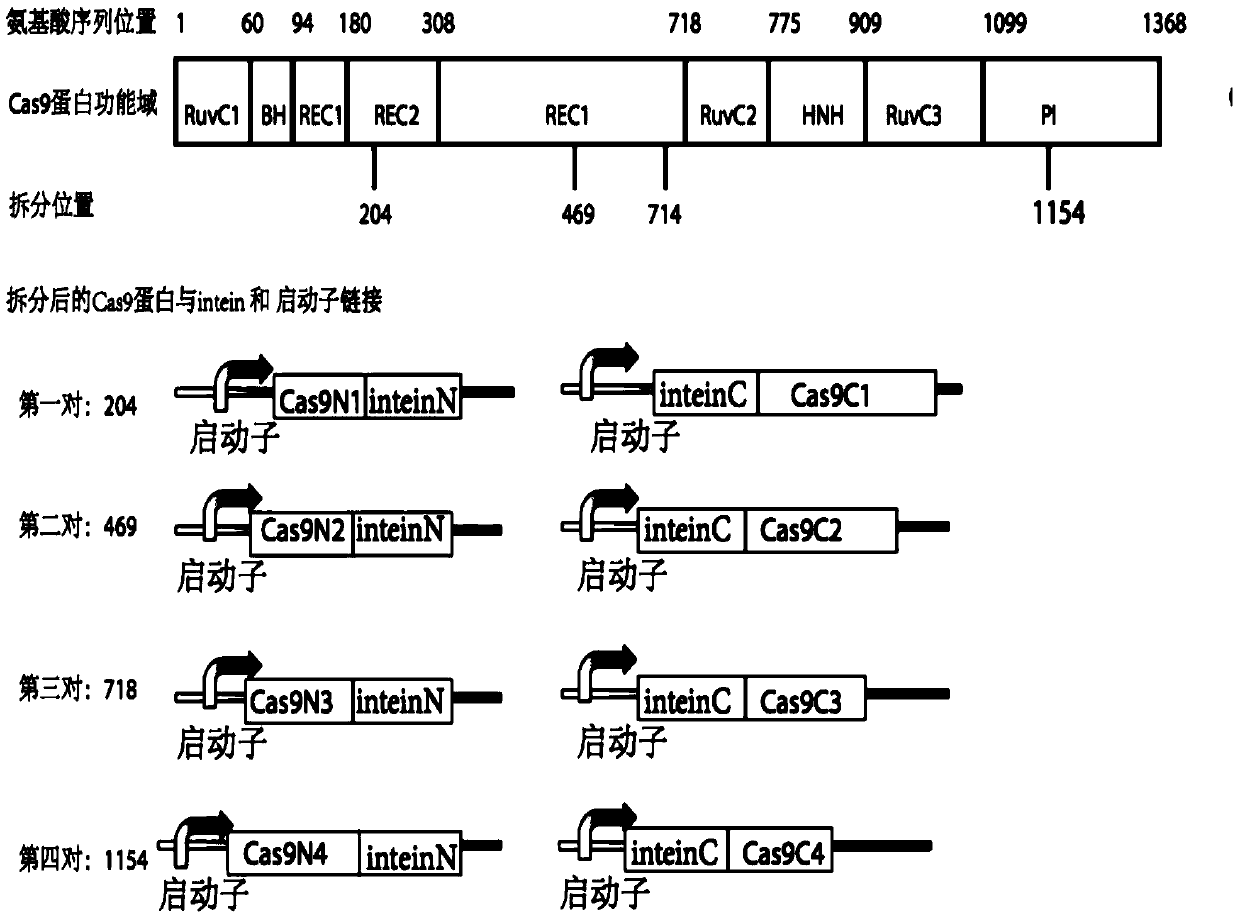
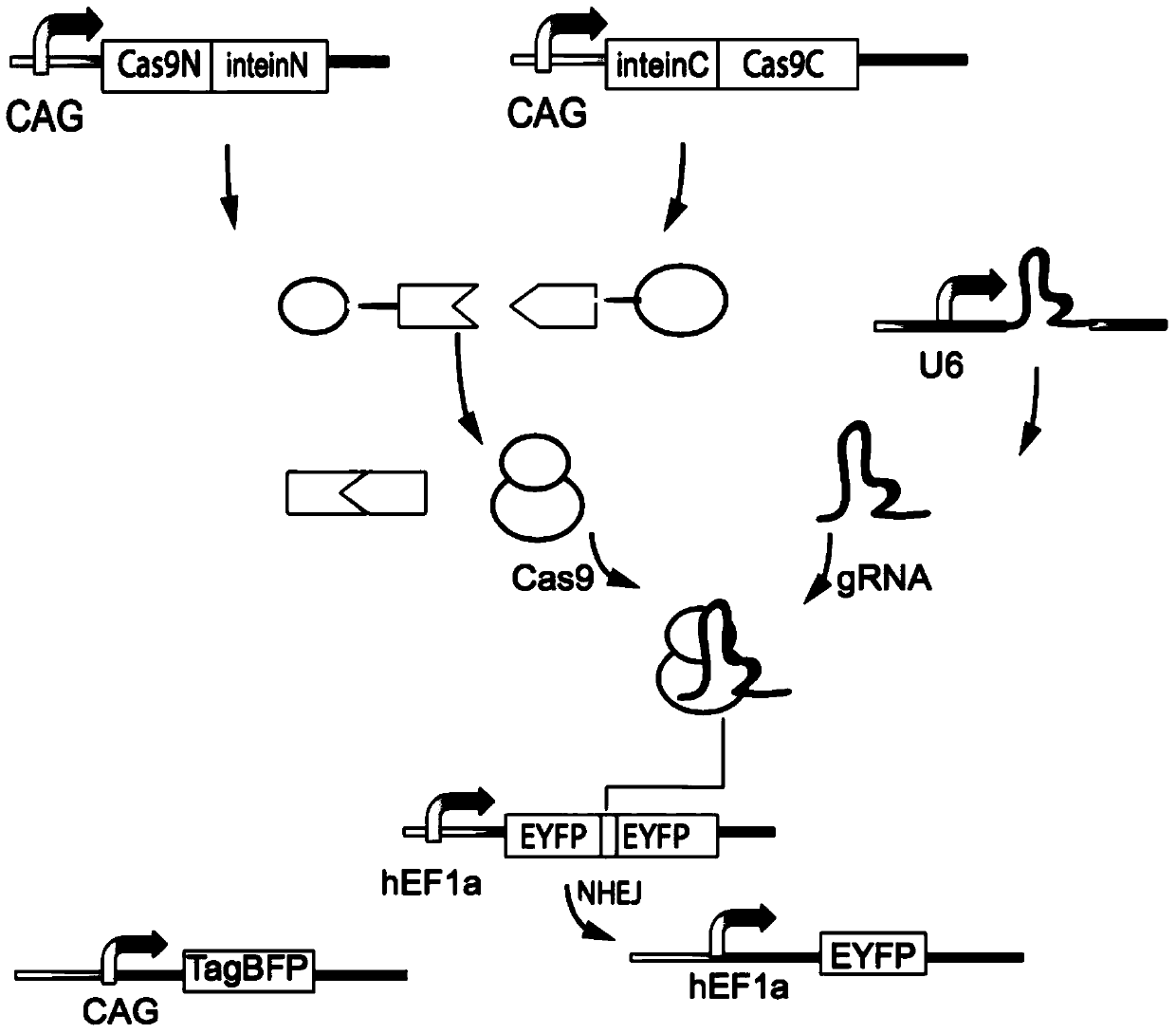
[0112] Using the split feature of the functional scope of Cas9 protein, the coding region of a Cas9 gene is split into N-terminal and C-terminal fragments (Cas9N and Cas9C), which are connected with the introns InteinN and InteinC, respectively. The Intein domain catalyzes protein cleavage, allowing it to recombine into a complete Cas9 protein in the cell for gene editing and gene expression regulation.
[0113] figure 1 It is a schematic diagram of the resolution of Cas9 protein, which is a different segment of amino acid sequence obtained by splitting the amino acid sequence of Cas9 protein from at least one of the following positions; the positions are between positions 203-204, positions 468-469, and positions 713-714 Between bits and 1153-1154.
[0114] The Cas9 protein used in the embodiment of the present invention is from Streptococcus pyogenes, namely hpCas9, a...
Embodiment 2
[0170] Example 2. Obtaining and functional verification of split dCas9 protein group and recombinant vector group
[0171] The split Cas9 proteins used in the following experiments are all dCas9 proteins and cannot be used for gene editing. They can only guide VP64 (amino acid sequence is sequence 9) or KRAB (amino acid sequence is sequence 10) for gene expression regulation. In addition, the separation site of the following experimental group is the position between amino acids 713-714 of the dCas9 protein.
[0172] The amino acid sequence of dCas9 protein is sequence 6 in the sequence listing, and the nucleotide sequence of its coding gene dCas9 is sequence 5 in the sequence listing.
[0173] The amino acid sequence of dCas9N is position 1-713 of sequence 6 in the sequence listing, and the nucleotide sequence of its coding gene dCas9N is position 1-2139 of sequence 5 in the sequence listing;
[0174] The amino acid sequence of dCas9C is at positions 714-1368 in sequence 6 in the seq...
Embodiment 3
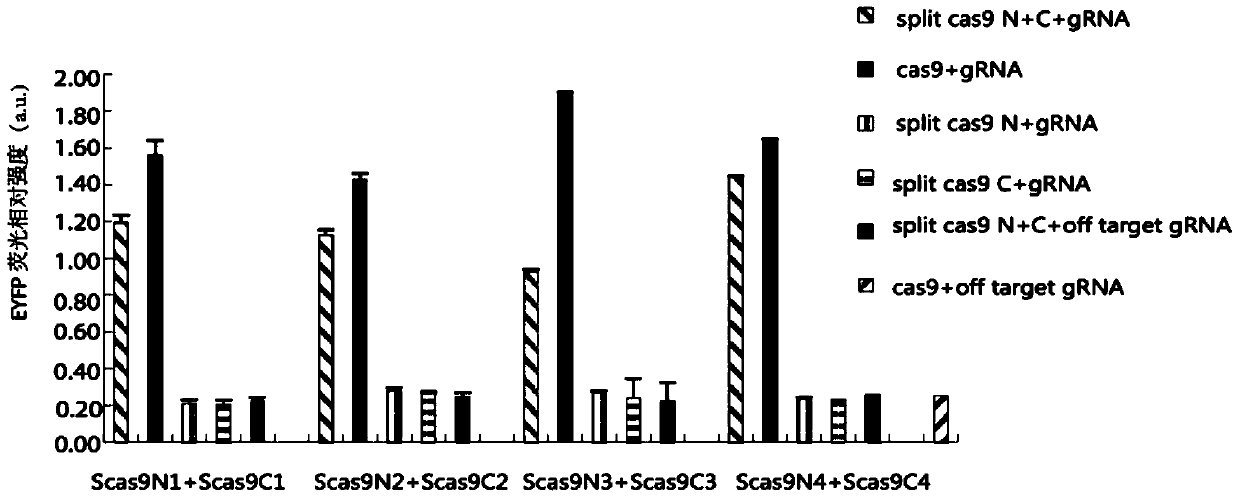
[0210] Example 3. Comparison does not use Intein-mediated split Cas9 system and Intein-mediated split Cas system for gene editing
[0211] In view of the article reports that the split Cas9 protein can be automatically recombined in the cell, in this implementation test, the 714th and 1154th amino acid positions of SpCas9 selected in the present invention were split for testing, and compared with Example 1.
[0212] 1. Whether the splitting of Spcas9 at position 714 uses Intein to mediate efficiency comparison
[0213] 1. Construction of recombinant vector
[0214] pCAG-SpCas9N714 plasmid, its nucleotide sequence is sequence 39, among which the 4635-6773th nucleotides of sequence 39 is the gene encoding protein SpCas9N714. The vector expresses protein SpCas9N714, and the amino acid sequence of this protein is sequence 2 1-713 .
[0215] pCAG-SpCas9C714 plasmid, the nucleotide sequence of which is sequence 40, the 4638-6599th nucleotides of sequence 40 is the gene encoding protein SpCas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com