A method of screening an HIV-1 activity determinative factor based on high-throughput RNAi
A technology of HIV-1, determinant, applied in the field of molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 Detection of HIV-1 LTR Regulated Transcriptional Activity Participated by Kinases
[0033] The HIV-1 LTR-hRluc / HSV TK-hLuc luciferase dual reporter plasmid was stably expressed in HEK293T by using shRNA library high-throughput RNAi screening technology for 474 kinases; Take the fluorescence intensity of hLuc (such as figure 1 shown in A);
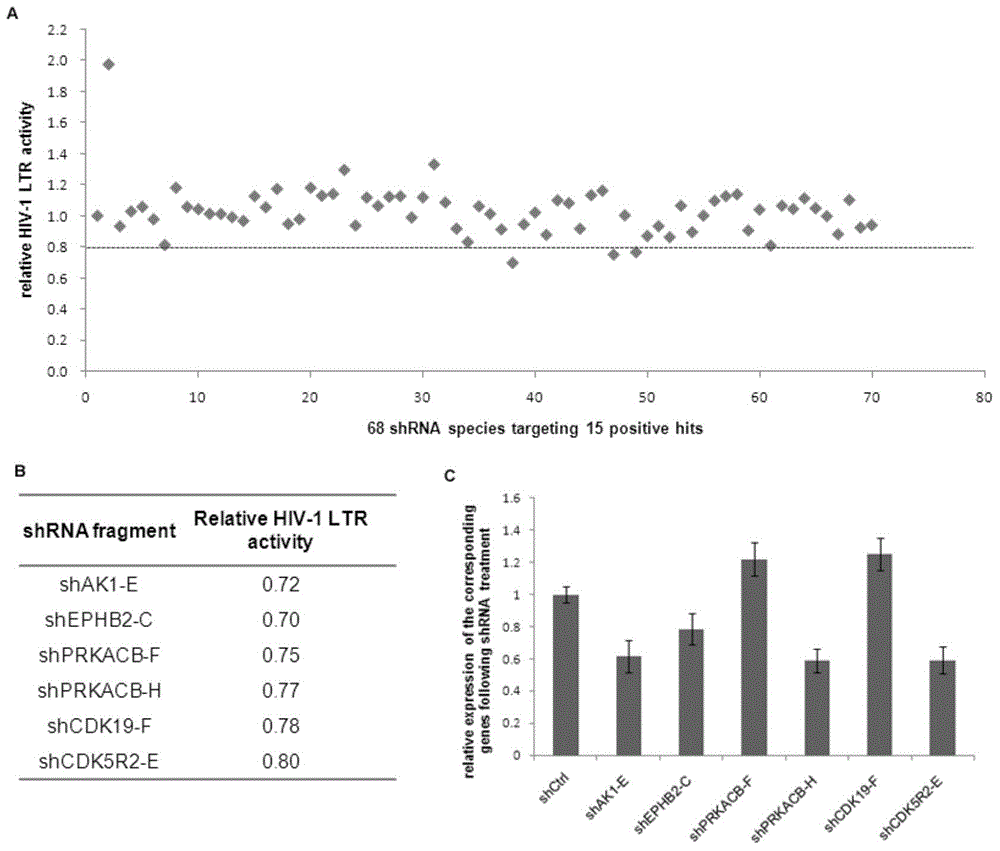
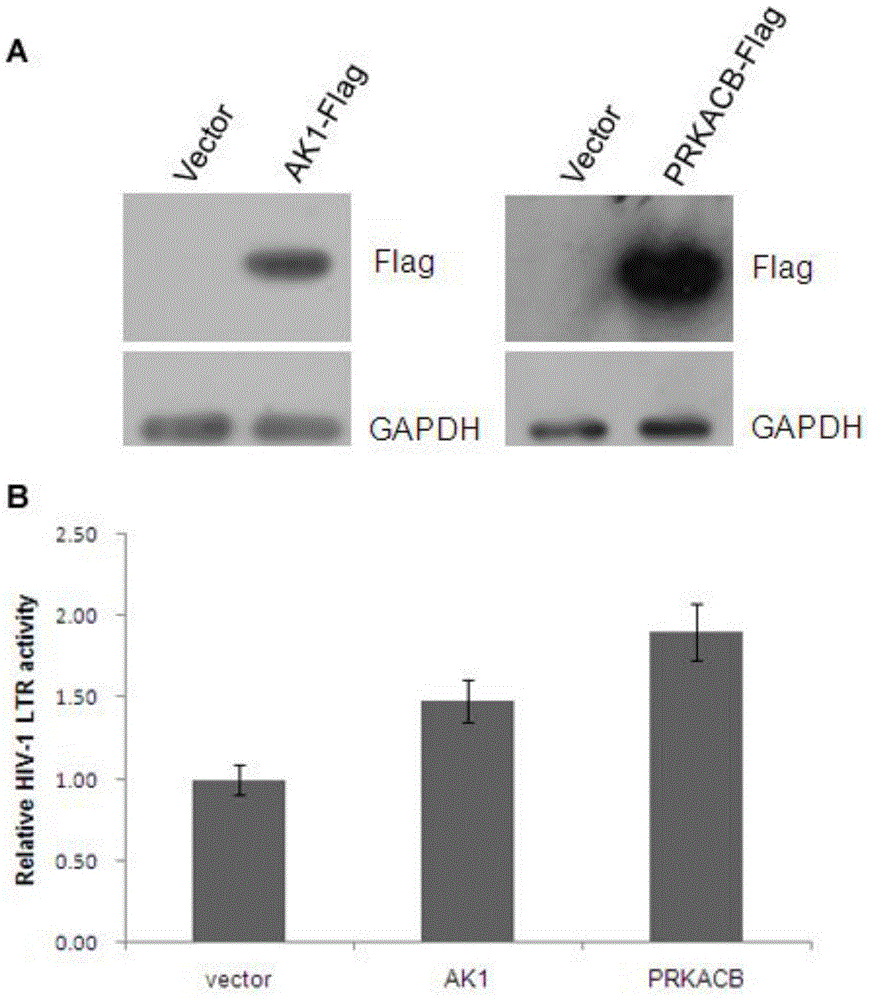
[0034] Two positive controls, NFκB and AKT1, were used for screening as shRNA or specific inhibitors (eg figure 1 Shown in B); inhibitors of NFκB or AKT1 kinase activity inhibited the activity of HIV-1 LTR by more than 30%, indicating that the screening system was reliable; in addition, the initial screening results showed 155 positive samples against HIV-1 LTR inhibited by 136 genes activity over 20% (eg figure 1 Shown in C), then above-mentioned 155 samples enter the second screening system, obtained 15 positive genes, the results are as shown in table 1;
[0035] Table 1. qPCR. Primers
[0036]
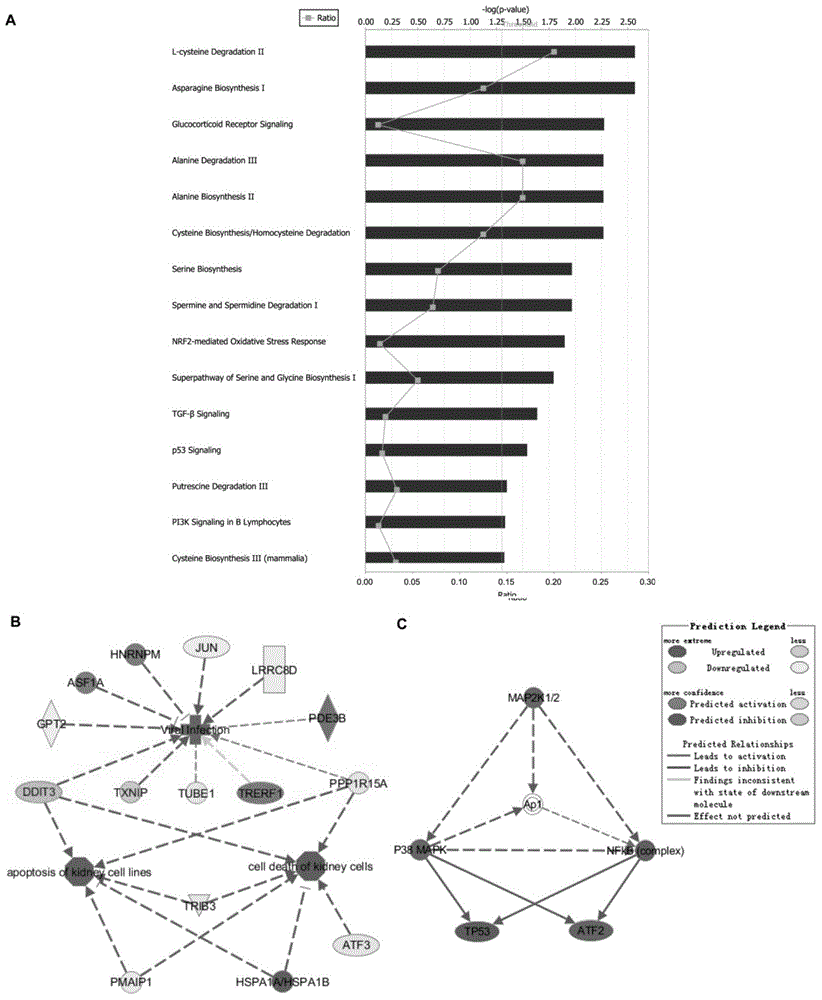
[0037] The resul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com