Method for separated identification and fine positioning of rice agronomic trait gene by whole genome sequencing
A Whole Genome Sequencing, Fine Mapping Technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0024] Materials and Methods:
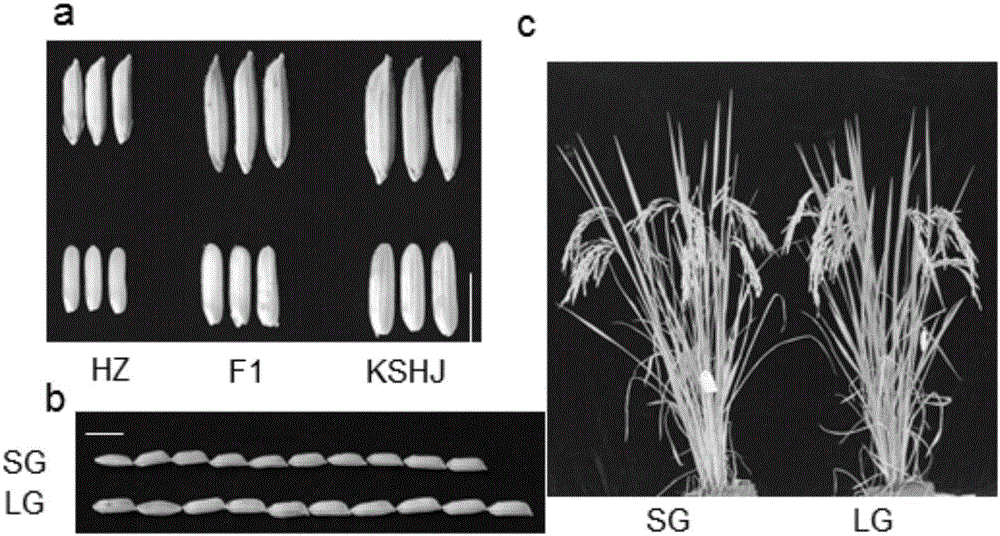
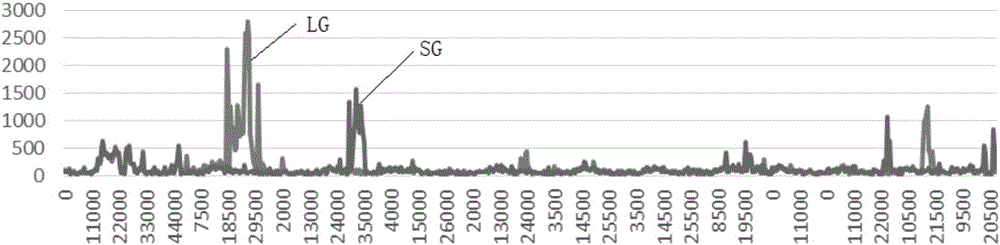
[0025] Rice material: Two groups of rice with different traits were used to validate the experimental method. One of them is the rice varieties Kuangsi Huaju (KSHJ, a large-grain indica variety) and Huazhan (HZ, a small-grain indica rice variety) with obvious differences in grain shape. Continuous backcrossing was carried out with Huazhan as the recurrent parent to obtain BC 4 f 1 A large single plant and a small single plant were selected from this group as sequencing samples; the other group consisted of the rice indica variety Fei B10 with long hairs on the leaf surface and the American rice variety Cypress with smooth leaves, and Cypress was used as the recurrent parent for continuous recurrent pay, get BC 4 f 1 Group, from which a single plant with hairy leaves on the surface and the recurrent parent Cypress were selected as sequencing samples. All the germplasm resources used in the present invention are provided by the germplasm resou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com