CAPs (C1eaved Amplified Polymorphic Sequences) molecular marker for identifying amaranthus tuberculatus based on SNP (Single Nucleotide Polymorphism) site and application thereof
A technology of molecular markers and amaranthus, which is applied in the fields of genetic engineering and molecular biology, can solve the problems of inability to further identify and achieve accurate detection and identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
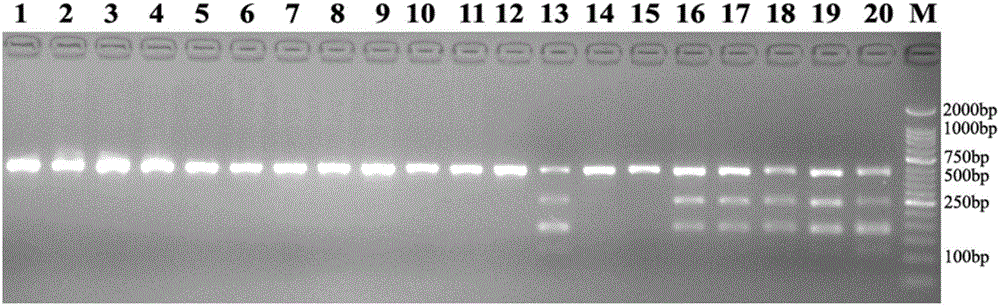
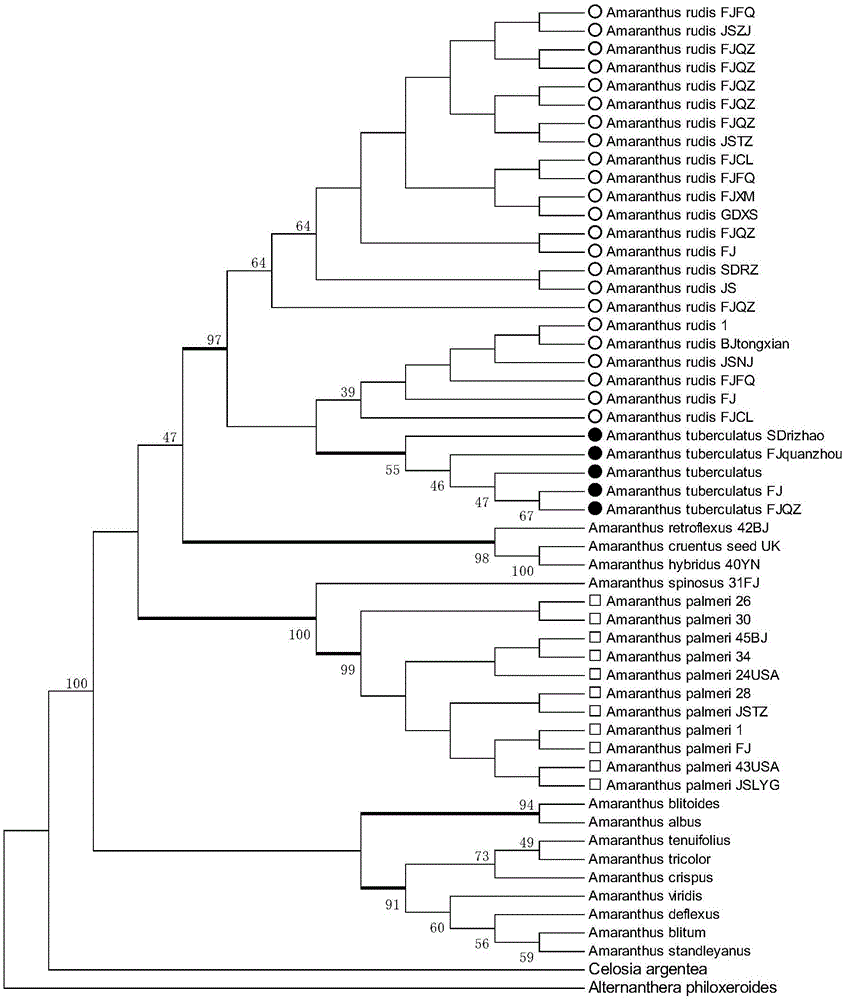
[0024] Example 1 is used to identify the acquisition of CAPs molecular markers of Amaranth
[0025] 1. Plant materials The specific names and sources of the materials selected in this example are shown in Table 1. The plant materials used in the experiment were leaves and seeds dried on silica gel.
[0026] Table 1 Plant materials and sources
[0027]
[0028]
[0029] 2. Reagents The reagents used for PCR amplification were purchased from Dalian Bao Biological Co., Ltd., the primers were synthesized by Beijing Aoke Dingsheng Biotechnology Co., Ltd., and the sequencing was completed by Shanghai Bioengineering Technology Service Co., Ltd.
[0030] 3. Method
[0031] 3.1 DNA extraction
[0032]Put 100mg of silica gel-dried plant leaves or seeds into a 2ml EP tube with 4mm steel balls added in advance, quickly freeze in liquid nitrogen for 30min, place the EP tube on a Geno / Grinder2000 (SPEX SamplePrep) high-throughput grinder, and grind for 1.5min. 1000rpm / min. Total l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com