Denovo sequencing method and device
A sequencing and node technology, applied in the field of bioinformatics, can solve problems such as difficult to distinguish similar peptide sequences, reduce the speed of peptide identification, etc., and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0038] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail below through specific embodiments in conjunction with the accompanying drawings. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
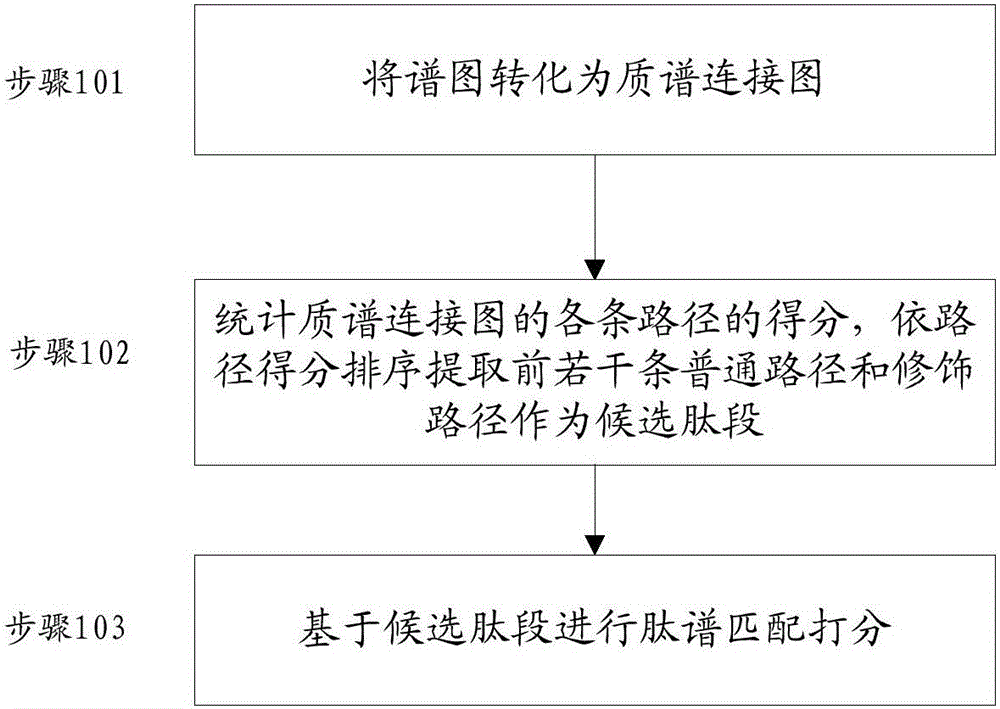
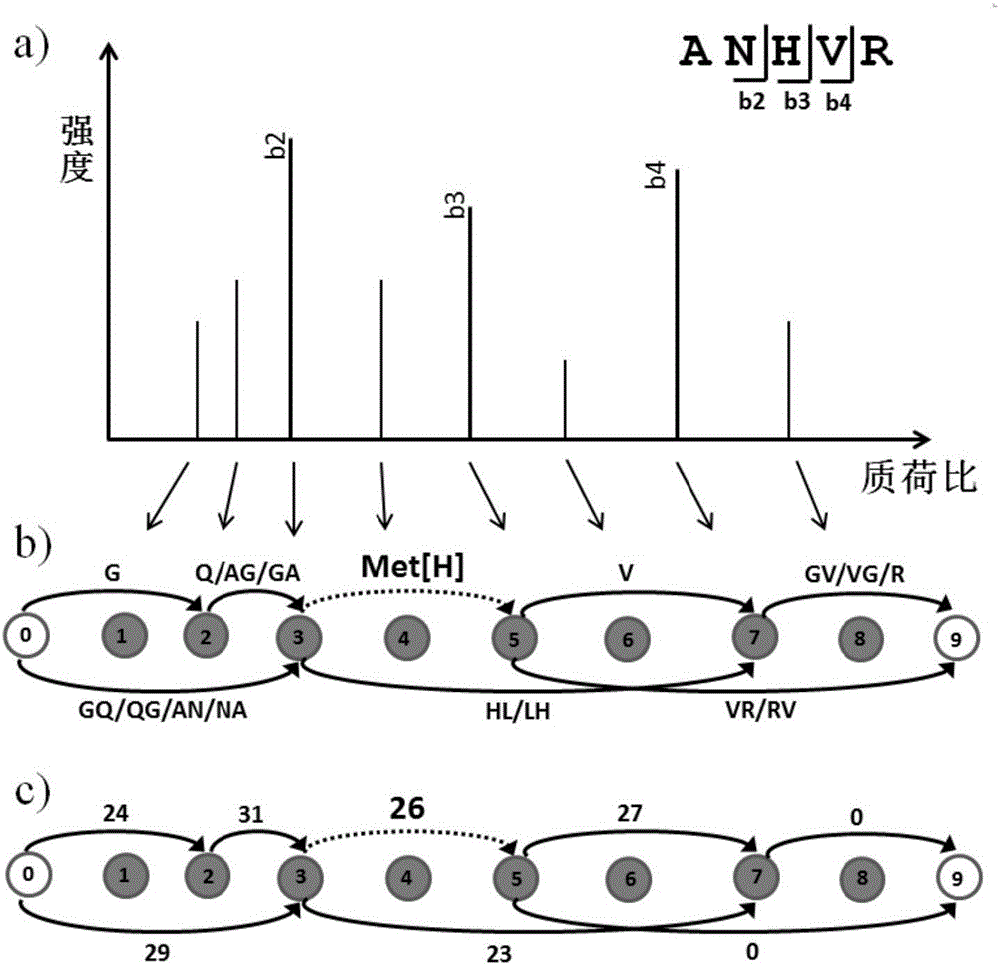
[0039] figure 1 A schematic flowchart of a de novo sequencing method according to an embodiment of the present invention is given. The method mainly includes converting the spectrogram into a mass spectrometry connection map (step 101); counting the scores of each path of the mass spectrometry connection map, sorting the first several common paths and modified paths according to the path scores as candidate peptides (step 102) and performing peptide spectrum matching scoring based on candidate peptides (step 103). Preferably, before performing the above steps, the received spectrogra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com