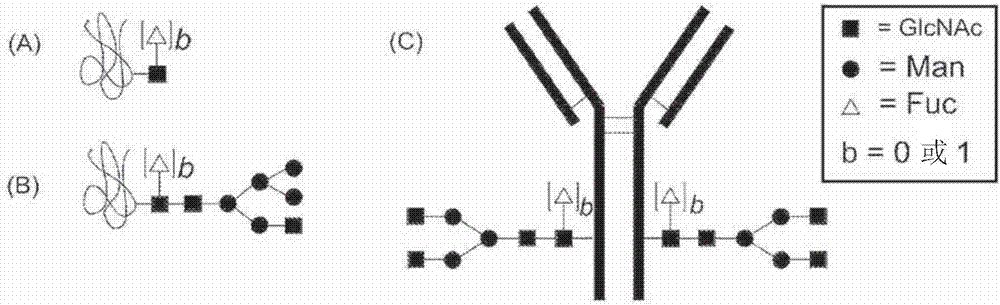
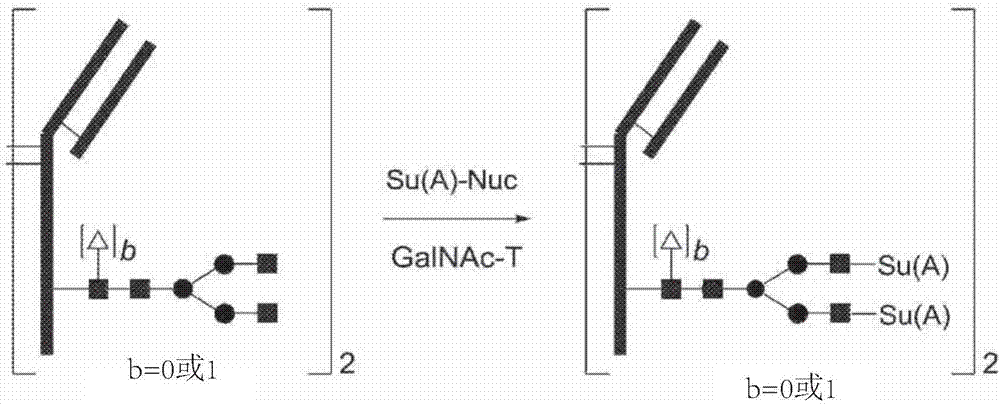
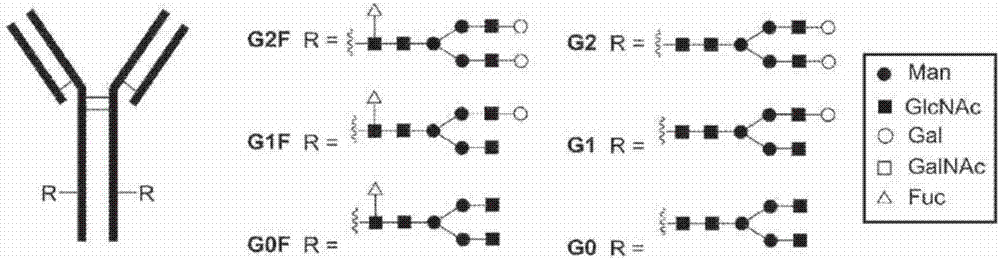
Process for modification of glycoprotein using Beta-(1,4)-n-acetylgalactosaminyl transferase or mutant thereof
A technology of acetylgalactosamine and glycoprotein, applied in the field of enzymatic modification of glycoprotein
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0392] Selection and design of embodiment 1.GalNAc transferase
[0393] Five specific sequences were selected for initial evaluation, specifically the Uniprot accession numbers: Q9GUM2 (C. elegans; represented herein as SEQ ID NO:2), U1MEV9 (Ascaris suum; represented herein as SEQ ID NO:3) , Q6J4T9 (Telina, represented herein as SEQ ID NO: 4), Q7KN92 (Drosophila melanogaster; represented herein as SEQ ID NO: 5), and Q6L9W6 (Homo sapiens).
[0394] The following polypeptides were constructed based on the predicted deletion of the cytoplasmic and transmembrane domains. These peptides contain predicted:
[0395] Caenorhabditis elegans (CeGalNAcT [30-383] represented by SEQ ID NO: 6)
[0396] KIPSLYENLTIGSSTLIADVDAMEAVLGNTASTSDDLLDTWNSTFSPISEVNQTSFMEDIRPILFPDNQTLQFCNQTPPHLVGPIRVFLDEPDFKTLEKIYPDTHAGGHGMPKDCVARHRVAIIVPYRDREAHLRIMLHNLHSLLAKQQLDYAIFIVEQVANQTFNRGKLMNVGYDVASRLYPWQCFIFHDVDLLPEDDRNLYTCPIQPRHMSVAIDKFNYKLPYSAIFGGISALTKDHLKKINGFSNDFWGWGGEDDDLATRTSMAGLKVSRYPTQIARYKMIKHSTEATNP...
Embodiment 2
[0406] Example 2. Design of the Caenorhabditis elegans GalNAcT mutant of I257
[0407] Based on the sequence alignment of CeGalNAcT and GalT, the isoleucine 257 Three active site mutants were designed with mutations to leucine, methionine or alanine (underlined).
[0408] CeGalNacT(30-383; I257L) represented by SEQ ID NO: 10
[0409] KIPSLYENLTIGSSTLIADVDAMEAVLGNTASTSDDLDTWNSTFSPISEVNQTSFMEDIRPILFPDNQTLQFCNQTPPHLVGPIRVFLDEPDFKTLEKIYPDTHAGGHGMPKDCVARHRVAIIVPYRDREAHLRIMLHNLHSLLAKQQLDYAIFIVEQVANQTFNRGKLMVASRLYPWQCFIFHDDVDLPEDDRNLK L FGGISALTKDHLKKINGFSNDFWGWGGEDDDLATRTSMAGLKVSRYPTQLARYKMIKHSTEATNPVNKCRYKIMGQTKRRWTRDGLSNLKYKLVNLELKPLYTRAVVDLLEKDCRRELRRDFPTCF
[0410] CeGalNAcT(30-383; I257M) represented by SEQ ID NO: 11
[0411] KIPSLYENLTIGSSTLIADVDAMEAVLGNTASTSDDLDTWNSTFSPISEVNQTSFMEDIRPILFPDNQTLQFCNQTPPHLVGPIRVFLDEPDFKTLEKIYPDTHAGGHGMPKDCVARHRVAIIVPYRDREAHLRIMLHNLHSLLAKQQLDYAIFIVEQVANQTFNRGKLMVASRLYPWQCFIFHDDVDLPEDDRNLK M FGGISALTKDHLKKINGFSNDFWGWGGEDDDLATRTSMAGLKVSRYPTQIAR...
Embodiment 3
[0414] Example 3. Design of the Caenorhabditis elegans GalNAcT mutant of M312
[0415] by adding methionine 312 CeGalNAcT mutants were designed by mutation to histidine.
[0416] CeGalNacT (30-383; M312H) represented by SEQ ID NO: 13
[0417] KIPSLYENLTIGSSTLIADVDAMEAVLGNTASTSDDLLDTWNSTFSPISEVNQTSFMEDIRPILFPDNQTLQFCNQTPPHLVGPIRVFLDEPDFKTLEKIYPDTHAGGHGMPKDCVARHRVAIIVPYRDREAHLRIMLHNLHSLLAKQQLDYAIFIVEQVANQTFNRGKLMNVGYDVASRLYPWQCFIFHDVDLLPEDDRNLYTCPIQPRHMSVAIDKFNYKLPYSAIFGGISALTKDHLKKINGFSNDFWGWGGEDDDLATRTSMAGLKVSRYPTQIARYK H IKHSTEATNPVNKCRYKIMGQTKRRWTRDGLSNLKYKLVNLELKPLYTRAVVDLLEKDCRRELRRDFPTCF
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com