Method for using computer program to simulate and generate simplified DNA methylation sequencing data
A computer program and sequencing data technology, applied in computing, electrical digital data processing, special data processing applications, etc., can solve problems that cannot be used to evaluate the reliability of splicing tools
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
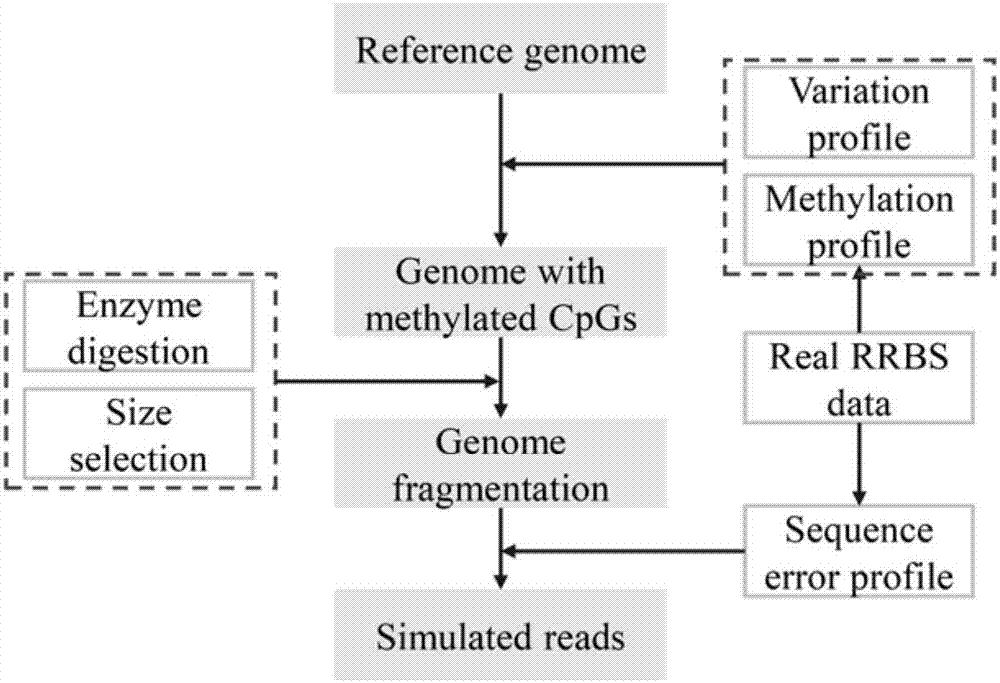
[0018] Embodiment 1: The method for generating simplified DNA methylation sequencing data by computer program (Python programming language) simulation provided by the present invention, first according to figure 1 The simulation shown produces simplified DNA methylation sequencing data:
[0019] (1) Simulation generates a reference genome sequence including single base insertion, deletion, single nucleotide variation and structural variation (these variation parameters can be given by the user), such as hg19.
[0020] (2) Simulating the methylation level at the CpG dinucleotide site on the reference genome obtained in step (1). Since the methylation level of CpG sites on the genome usually obeys the Beta distribution, we use the Beta model to generate methylation level values on CpG sites. In addition, considering the strong correlation of adjacent CpG sites in real data, we corrected the methylation levels of CpG sites within a distance of 100 bp based on a maximum likelih...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com