Method for efficiently detecting EGFR T790M mutant, probe and kit for detection
An EGFRT790M, mutant technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problem of low ctDNA fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
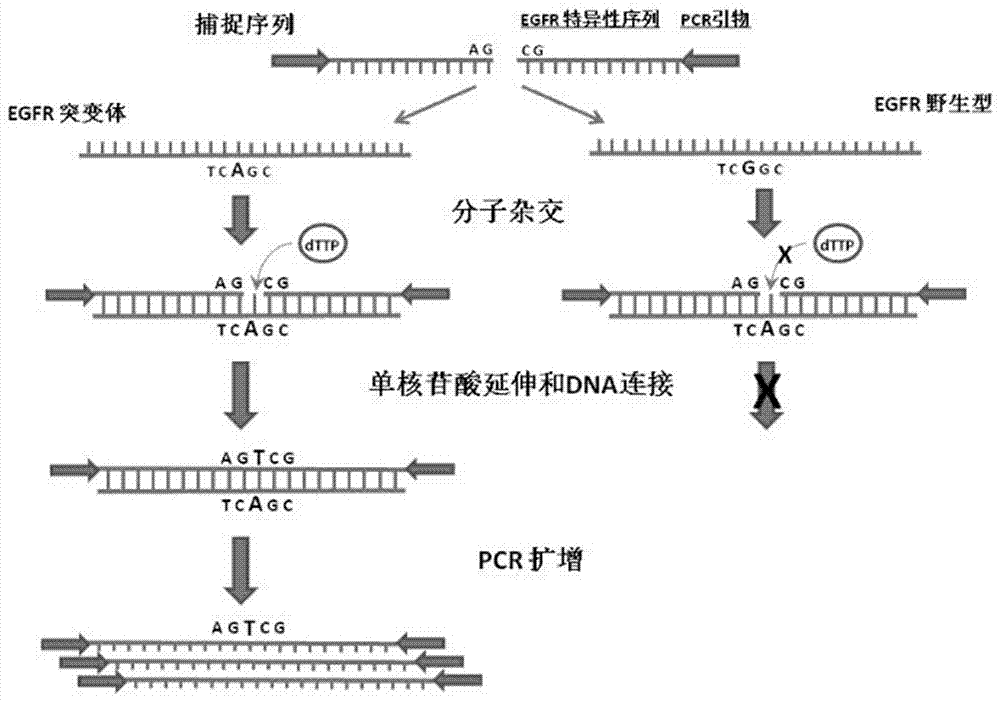
Image
Examples
Embodiment 1
[0036] Example 1 Mutant capture and real-time quantitative PCR detection
[0037] Materials used:
[0038] 2×EGFR mutant detection solution: 0.2% TritonX-100; 10-20mM MgCl 2 ; 16mM (NH4) 2 SO4; 50mM KCl; 10mM NAD, 40mM Tris-HCl (pH8.0); 0.6M each of the upstream and downstream mutant capture primers, blocking sequence 6M, dTTP 500M, HotTaq enzyme 1.0-3.0U, Ampligase 2.0-5.0U .
[0039] 10x PCR primer and probe solution: PCR upstream and downstream primers 3M each, four dNTPs 5mM each, mutant probe 0.3mM, internal control upstream and downstream primers 3M, internal control probe 0.3mM. (EGFR Exon 25)
[0040] SEQ ID NO.5:
[0041] Upstream mutant capture primer: 5' tcataccgtcattcatgc ctcacctccaccgtgcarctcatca 3';
[0042] SED ID NO.6:
[0043] Downstream mutant capture primer: 5' gcagctcatgcccttcggctgcctc acgattccacgaccgatg 3';
[0044] SEQ ID NO.4:
[0045] Mutant probe: 5'-tga gct gC a Tga tga gyt-3'; the 8th and 10th bases from the 5' end of the sequence are lock...
Embodiment 2
[0073] Embodiment 2 mutant capture and digital PCR detection
[0074] Materials used:
[0075] 2×EGFR mutant detection solution: 0.2% TritonX-100; 10-20mM MgCl 2 ; 16mM (NH4) 2 SO4; 50mM KCl; 10mM NAD, 40mM Tris-HCl (pH8.0); 0.6M each of the upstream and downstream mutant capture primers, blocking sequence 6M, dTTP 500M, HotTaq enzyme 1.0-3.0U, Ampligase 2.0-5.0U .
[0076] 10x PCR primer and probe solution: PCR upstream and downstream primers 3M each, four dNTPs 5mM each, mutant probe 0.3M, internal control upstream and downstream primers 3M, internal control probe 0.3M. (EGFR Exon 25)
[0077] SEQ ID NO.5:
[0078] Upstream mutant capture primer: 5' tcataccgtcattcatgc ctcacctccaccgtgcarctcatca 3';
[0079] SEQ ID NO.6:
[0080] Downstream mutant capture primer: 5' gcagctcatgcccttcggctgcctc acgattccacgaccgatg 3';
[0081] SEQ ID NO.4:
[0082] Mutant probe: 5'-tga gct gC a Tga tga gyt-3', the 8th and 10th bases from the 5' end of the sequence are locked nucleic aci...
Embodiment 3
[0106] Example three mutant capture and conventional PCR detection
[0107] Materials used:
[0108] 2×EGFR mutant detection solution: 0.2% TritonX-100; 10-20mM MgCl 2 ; 16mM (NH4) 2 SO4; 50mM KCl; 10mM NAD, 40mM Tris-HCl (pH8.0); contains 0.6M upstream and downstream mutant capture primers, blocking sequence 6M, dTTP 500mM, HotTaq enzyme 1.0-3.0U, Ampligase 2.0-5.0U .
[0109] 10x PCR primer solution: 3M each of PCR upstream and downstream primers, 5mM each of the four dNTPs.
[0110] SEQ ID NO.5:
[0111] Upstream mutant capture primer: 5' tcataccgtcattcatgc ctcacctccaccgtgcarctcatca 3';
[0112] SEQ ID NO.6:
[0113] Downstream mutant capture primer: 5' gcagctcatgcccttcggctgcctc acgattccacgaccgatg 3';
[0114] SEQ ID NO.4:
[0115] Mutant probe: 5'-tga gct gCa Tga tga gyt-3', the 8th and 10th bases from the 5' end of the sequence are locked nucleic acid modified bases, and the 9th base from the 5' end is 2 -Aminodeoxyadenylic acid modified base, the 5' end is conne...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com