Process for detection of DNA modifications and protein binding by single molecule manipulation
A molecular, protein-based technology used in the detection of DNA modifications and protein binding by single-molecule manipulation, which can solve problems such as the lack of insights into genome-wide biological functions, improve statistics, reduce instrument drift, and eliminate false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0195] Experimental example
Background of the invention
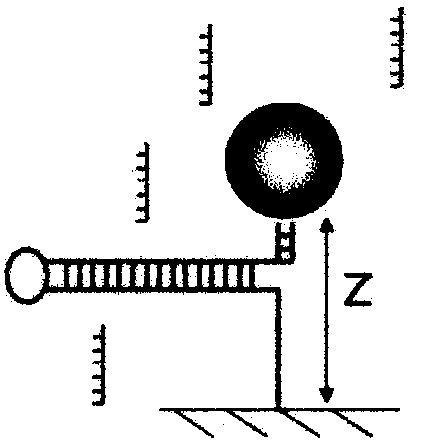
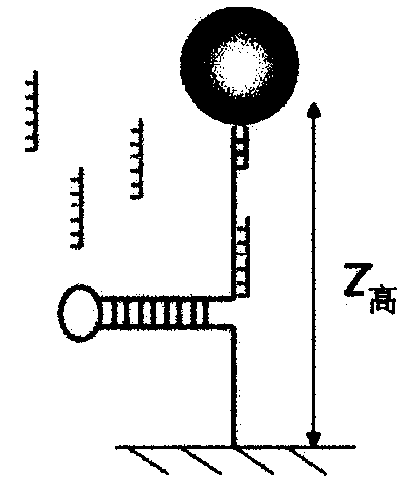
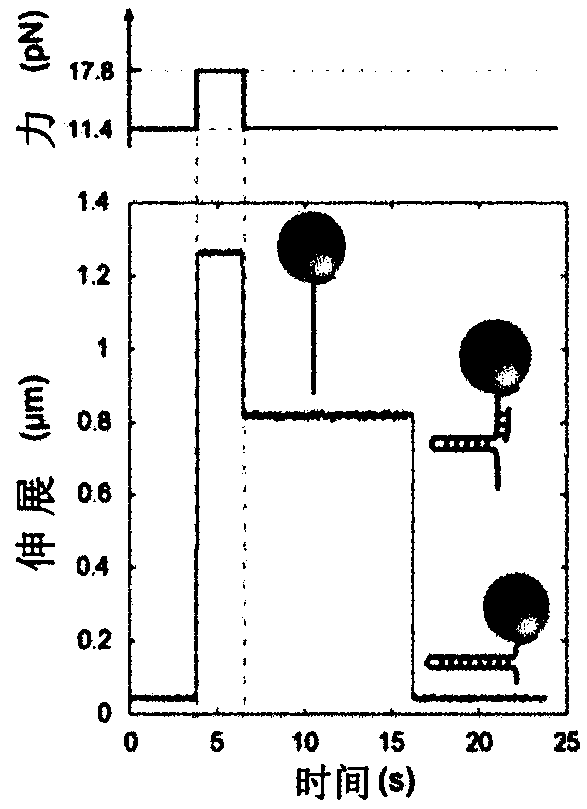
[0196] Binding of proteins to DNA is an important phenomenon in biology; it is a very general process that controls many reactions. While the thermodynamic equilibrium nature of this mechanism is well known, measuring its kinetics is a more challenging problem. The use of single molecules not only provides the ability to measure the time it takes a protein to find its DNA target but also the exact location of a binding event. We describe here a new single-molecule assay that achieves these goals.
[0197] Although this assay is broad in scope, we first demonstrate its applicability to binding of specific oligonucleotides, as well as non-specific binding of helicases to ssDNA.
[0198] Finally, we discuss the specific binding of antibodies that recognize methylated sites in DNA.
[0199] Summary
[0200] The present invention relates to novel methods for detecting a wide variety of DNA modification and DNA protei...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com