Kit for detecting methylation of promoter region of fragile X mental retardation 1 (FMR1) gene through fluorescent quantitative method and application thereof
A technology of fluorescence quantification and kit, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc. It can solve the problems of poor specificity and difficulty in screening a large number of samples, and achieve the effect of reducing non-specificity and pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
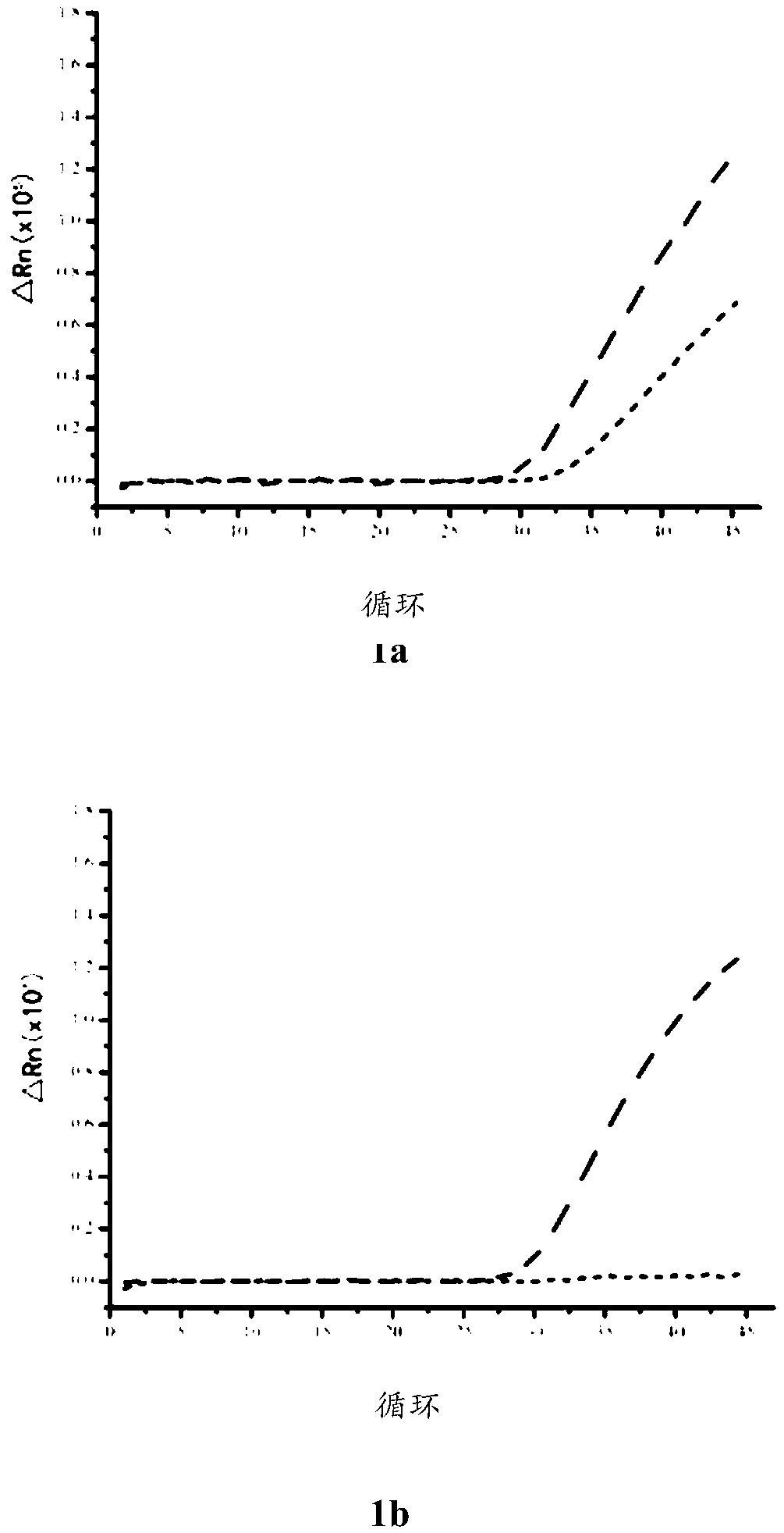
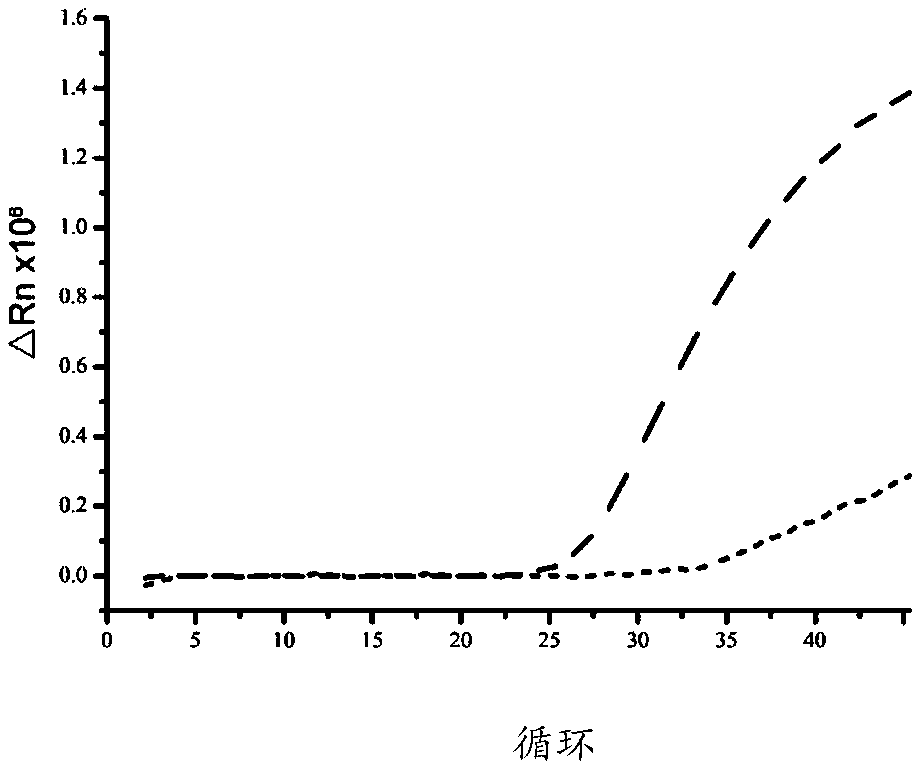
Image
Examples
Embodiment 1
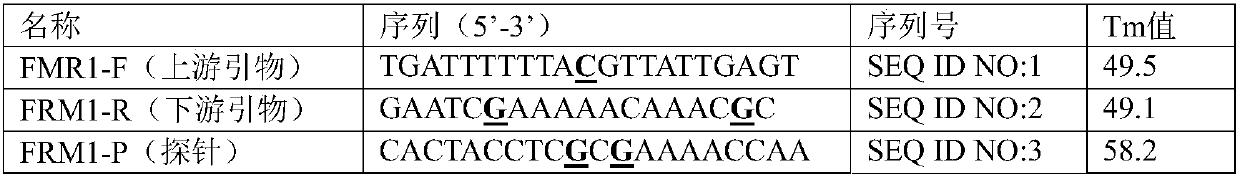
[0015] Example 1. Primers and probes for detecting methylation of FMR1 gene by qPCR method
[0016] In order to overcome the above-mentioned defects in the MS-PCR method, the present invention provides a kit for detecting the methylation of the promoter region of the FMR1 gene based on a fluorescence quantitative method. The sequences of primers and probes involved in the present invention are listed in Table 1 below.
[0017] Table 1. Primers and probes for detecting methylation of FMR1 gene by qPCR method
[0018]
[0019] The underlined and bold bases are designed for the methylation sites of the FMR1 gene, and the size of the qPCR product is 143bp. The probe has a fluorescent reporter group FAM at its 5' end and a fluorescent quencher group MGB at its 3' end.
[0020] in the CG of the upstream primer in the GC in the downstream primer Both target methylated sequences that remain after sulfite treatment or The characteristics of the design; if no methylation oc...
Embodiment 2
[0025] Embodiment two. the application of kit of the present invention
[0026] The type of sample detected by the present invention is a clinical sample of peripheral whole blood. After the genomic DNA is extracted from the blood sample, PCR+capillary electrophoresis method is used to confirm that the tested sample method is Fragile X positive and Fragile X negative.
[0027] The extracted DNA was treated with a sulfite treatment kit of Jiangsu Jingshan Biotechnology Co., Ltd. in an amount of 400 ng to recover the converted DNA, and the eluted DNA was 60 μl. The reaction was carried out through the following fluorescent quantitative PCR reaction system. The qPCR reaction system used the product of Promega Company in the United States, as shown in Table 3, and the reaction conditions were shown in Table 4.
[0028] Table 3. Reaction system for detection of methylation in the promoter region of FMR1 gene by qPCR
[0029] name
Volume (20μl reaction system)
Fina...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com