DNA chip for detecting main drug-resistant mutation of HIV reverse transcriptase regions, preparation thereof and application thereof
A technology of reverse transcriptase region and DNA chip, which is applied in the direction of biochemical equipment and methods, microbial measurement/testing, etc., and can solve problems such as HIV drug-resistant mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1, design and preparation of biochip
[0027] (1) Primer and probe design
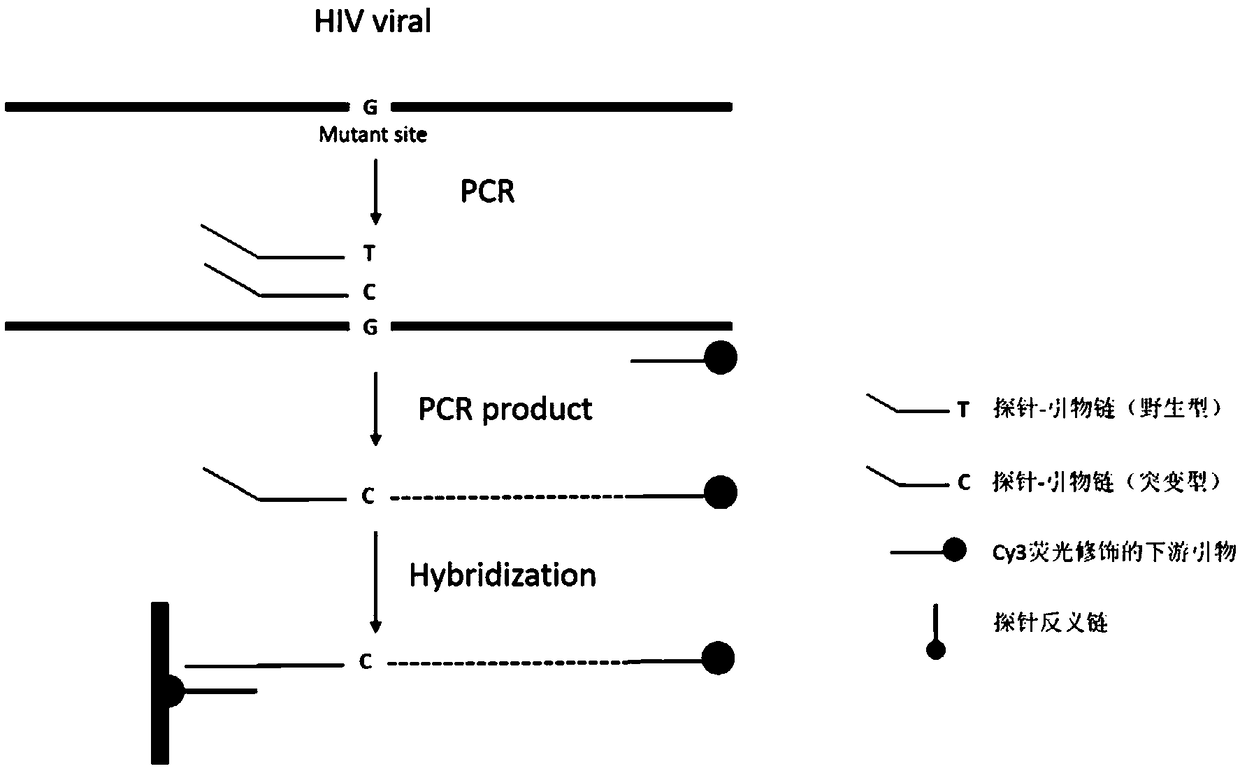
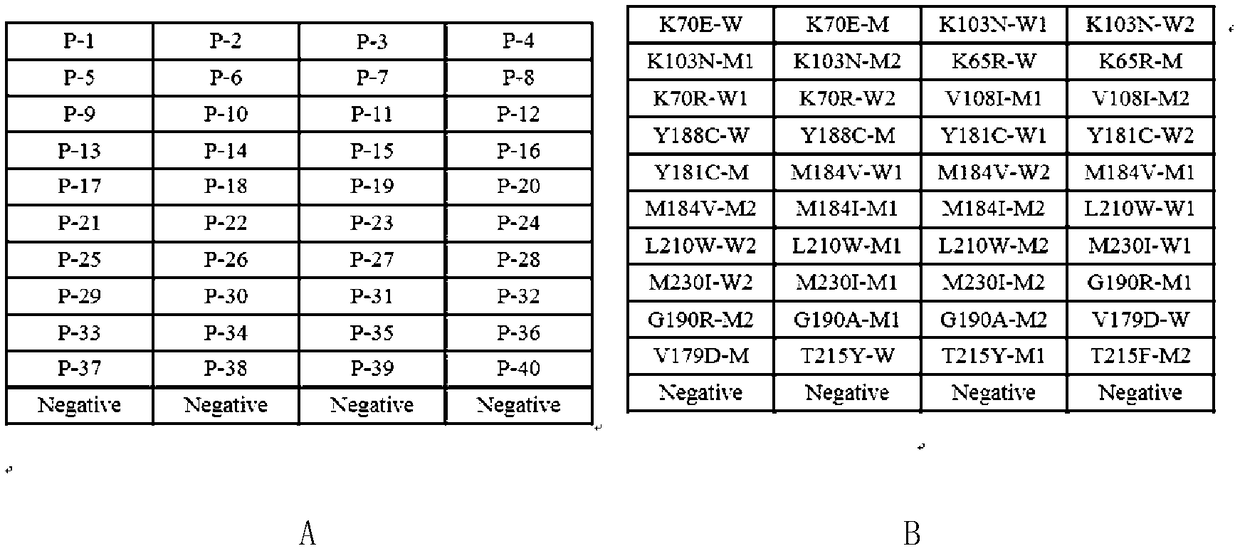
[0028] 16 drug-resistant mutation sites in the HIV reverse transcriptase region were used as detection genes, and the 16 common drug-resistant sites were: K70E, K103N, K65R, K70R, V108I, Y188C, Y181C, M184V, M184I, L210W, M230I, G190R . Designed according to the principle of allele-specific detection, the primers include a universal downstream primer and multiple specific upstream primers (18-27bp in length), in which the 3' terminal base of the wild-type specific upstream primer at the drug-resistant site is the same as the detected site The wild-type site sequence of the point is completely complementary, and the 3' terminal base of the mutant-type specific upstream primer is completely complementary to the mutant-type site sequence of the detected site, so when performing PCR detection, the specific upstream primer only amplifies the the complementary sequence. A general downstr...
Embodiment 2
[0043] Example 2, the application of biochips in the detection of drug-resistant mutations in the HIV reverse transcriptase region
[0044] (1) PCR amplification of the sample to be tested
[0045] The DNA or cDNA of the sample to be detected, the cy3-modified probe-primer chain, and the universal downstream primer are used for PCR reaction in the same tube.
[0046]The amplification template of conventional PCR amplification method is cDNA obtained by reverse transcription of RNA extracted from HIV patient plasma or DNA extracted from artificially constructed plasmid. The artificially constructed plasmids in this example are the recombinant plasmids containing the HIV reverse transcriptase region gene constructed with the pUCm-19 vector in the previous laboratory, including wild-type plasmids without drug-resistant mutations and mutant plasmids containing the above-mentioned drug-resistant mutations: Plasmid-HIV-WT, Plasmid-K70E, Plasmid-K103N, Plasmid-K65R, Plasmid-K70R, Pl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com