Amplification of target sequences
A technology of primer amplification and DNA sequence, which is applied in the field of target sequence amplification, and can solve the problems of the possibility of uneven sequence coverage, excessive or insufficient specific regions, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0355] Example 1-Whole genome amplification and gene targeting procedures
[0356] The following procedures are used for whole genome amplification (WGA) and gene targeting studies described herein.
[0357] 1. Materials and methods
[0358] The first round of whole genome amplification and gene targeting procedures:
[0359] (i) To 6.5μl PCR grade H 2 O add 1μl cell lyase (heat-resistant protease; EmbryoCellect TM Kit, RHS; number RHS1001); this is called cell lyase Dil 1.
[0360] (ii) Generate a cell lysis mixture by mixing the reagents in Table 1:
[0361] Table 1
[0362] Component
Volume for 1 cleavage reaction
PCR grade H 2 O
2.7
0.15
0.15
total capacity
3
[0363] (iii) Add 3 μl of cell lysis mix to the cells or DNA tube. Tap the solution to the bottom of the tube or centrifuge briefly. Don't vortex.
[0364] In this regard, a single cell usually contains about 6 pg of DNA.
[0365] (iv) Incubate the sample in a thermal cycler with th...
Embodiment 2
[0402] Example 2-Using a single cell to verify the enrichment of targeted genes in WGA PCR
[0403] Research is conducted to validate the protocol for enriching target genes.
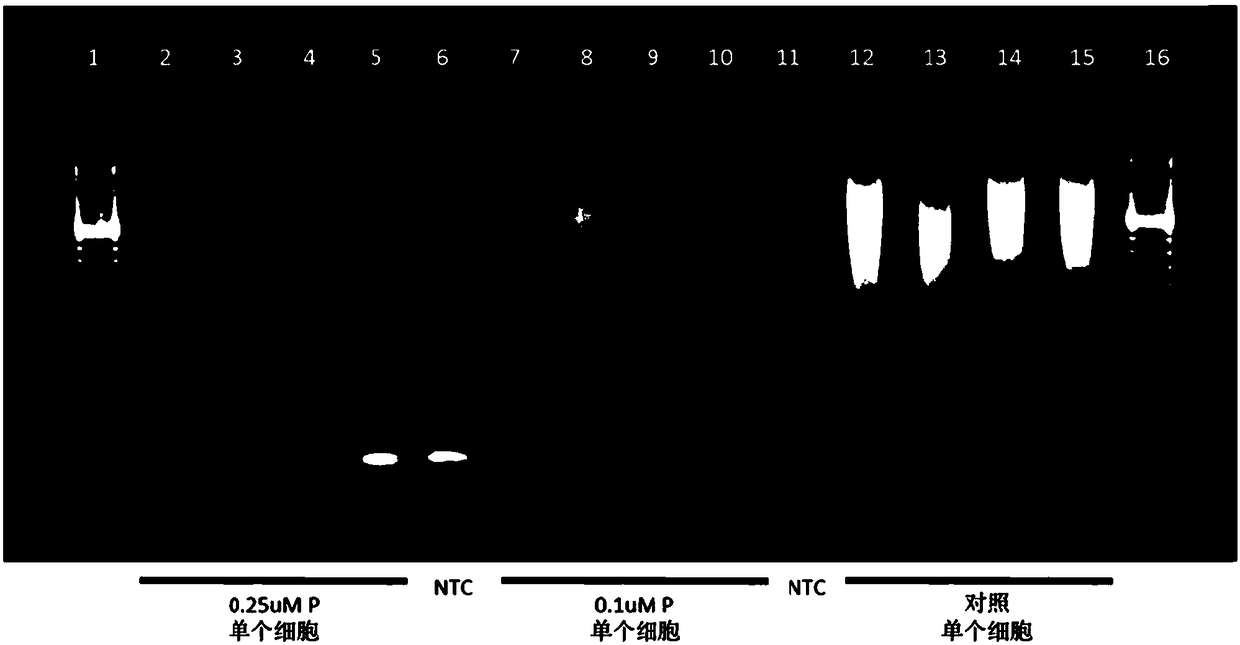
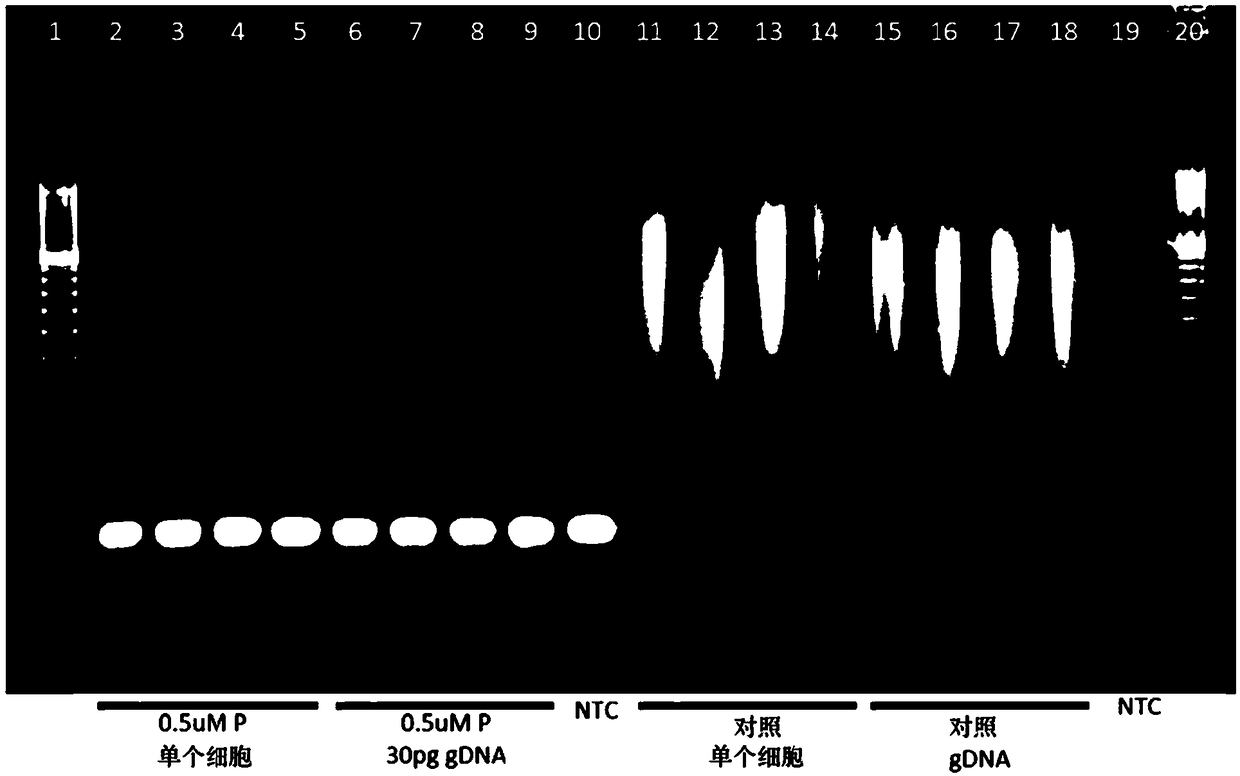
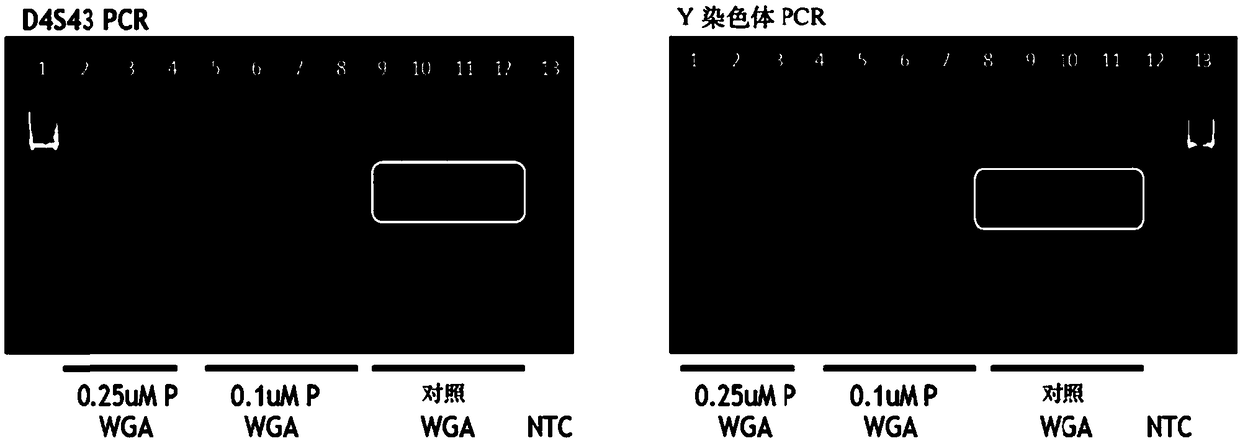
[0404] Add two sets of gene-specific primers (P) (0.5μM, 0.25μM or 0.1μM concentration) to WGA (multiplex PCR; figure 1 with figure 2 ), and a single cell manually sorted from a commercially available cell line (Coriell; 47, XY, +15 or 48, XXY, +21) was used for the first round of whole genome amplification and gene targeting as described in Example 1. Xiang (gene targeting).
[0405] As can be seen, the amplification process results in whole-genome amplification of the genomic DNA and specific amplification of the targeted gene, and amplification of the targeted gene is observed at the selected primer concentration tested. Note that the amplification of WGA in the presence of the gene-specific primer (P) is slightly weaker than the control WGA. These results confirm that overlapping WGA and gene-specific ...
Embodiment 3
[0413] Example 3-Targeting sequence enrichment (TSE) of HBB gene
[0414] In order to further confirm that this procedure can effectively amplify other target sequences, and to evaluate the use of this procedure in genetic screening and / or diagnosis, for example, this procedure is used for preimplantation genetic diagnosis and preimplantation genetic screening. This procedure is used to analyze the enrichment of the target sequence of the HBB gene encoding β globin.
[0415] Multi-cell samples (5-cells) were used to carry out combined whole-genome amplification and HBB gene targeted amplification using two primer sets. Then the PCR products were seeded into two specific HBB PCRs; mainly amplified exons 1 and 2, and amplified exon 3. The second round of HBB PCR amplicons were combined with whole-genome amplified DNA with targeted amplification of the HBB gene to generate a final dilution of 1:20. WGA-only PCR products, combined whole-genome amplification and HBB gene targeted amp...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com