Mature miRNA full-site recognition method based on SVM-AdaBoost
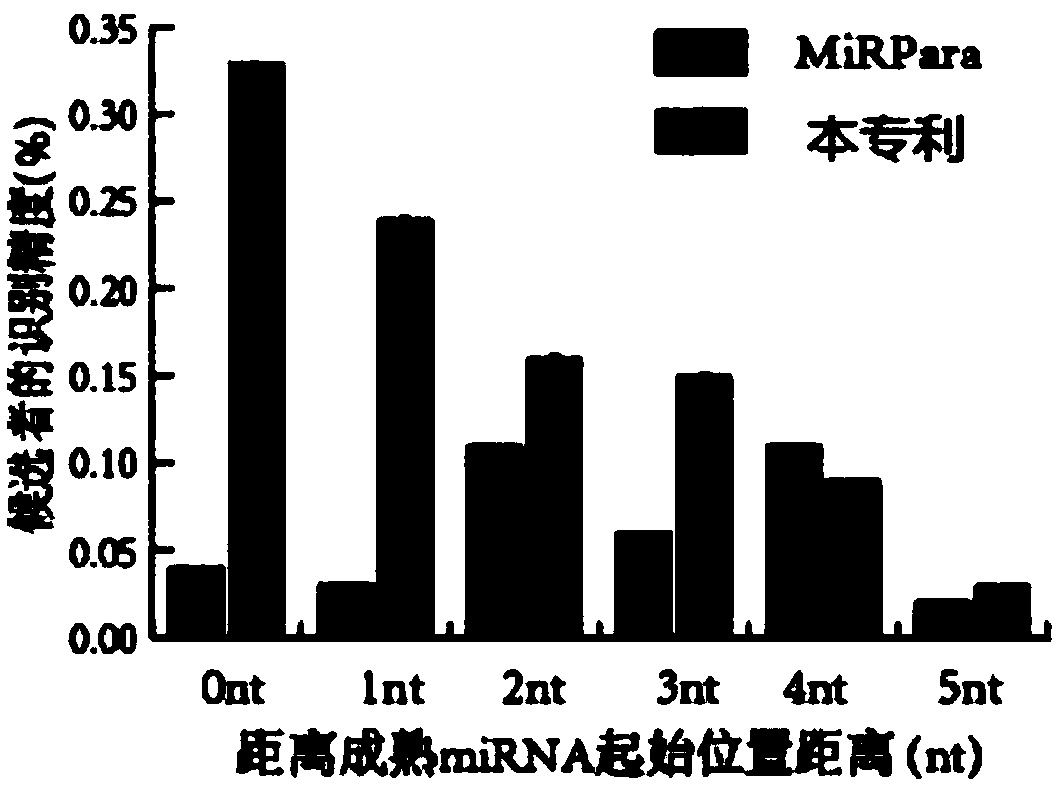
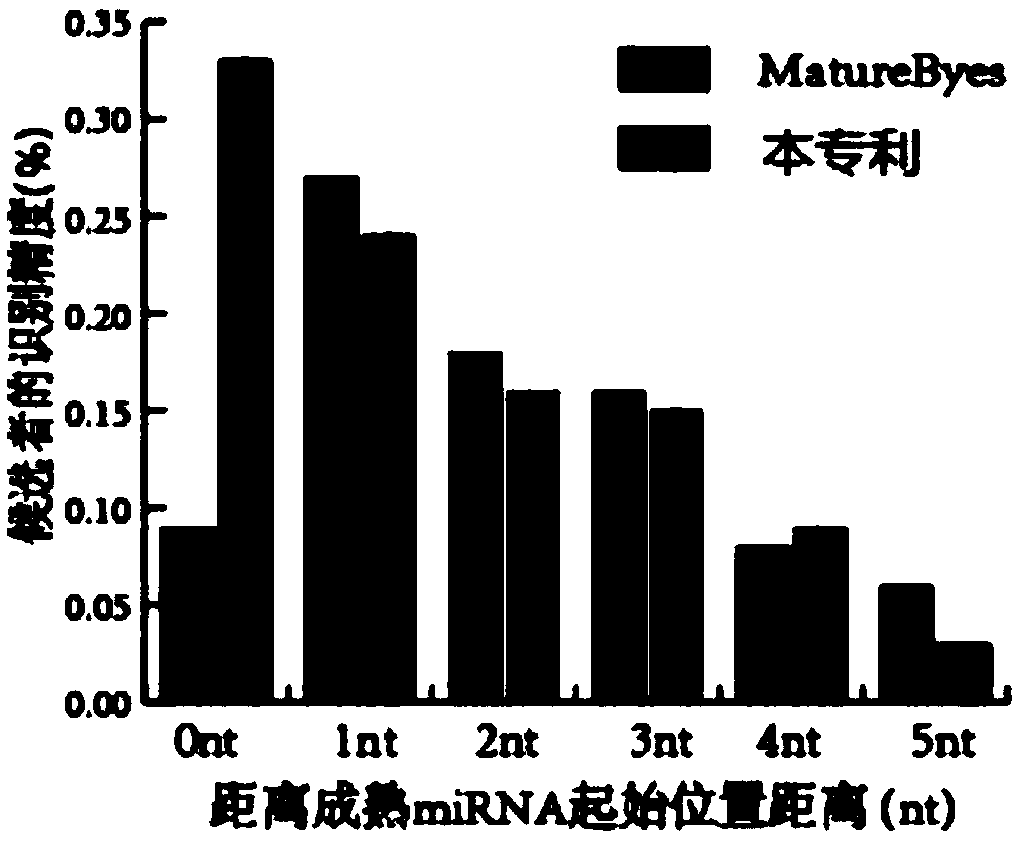
A recognition method and a mature technology, applied in the field of bioinformatics, can solve problems such as class imbalance and low precision, achieve high classification performance, improve recognition precision, and reduce the average number of nucleotide offsets
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
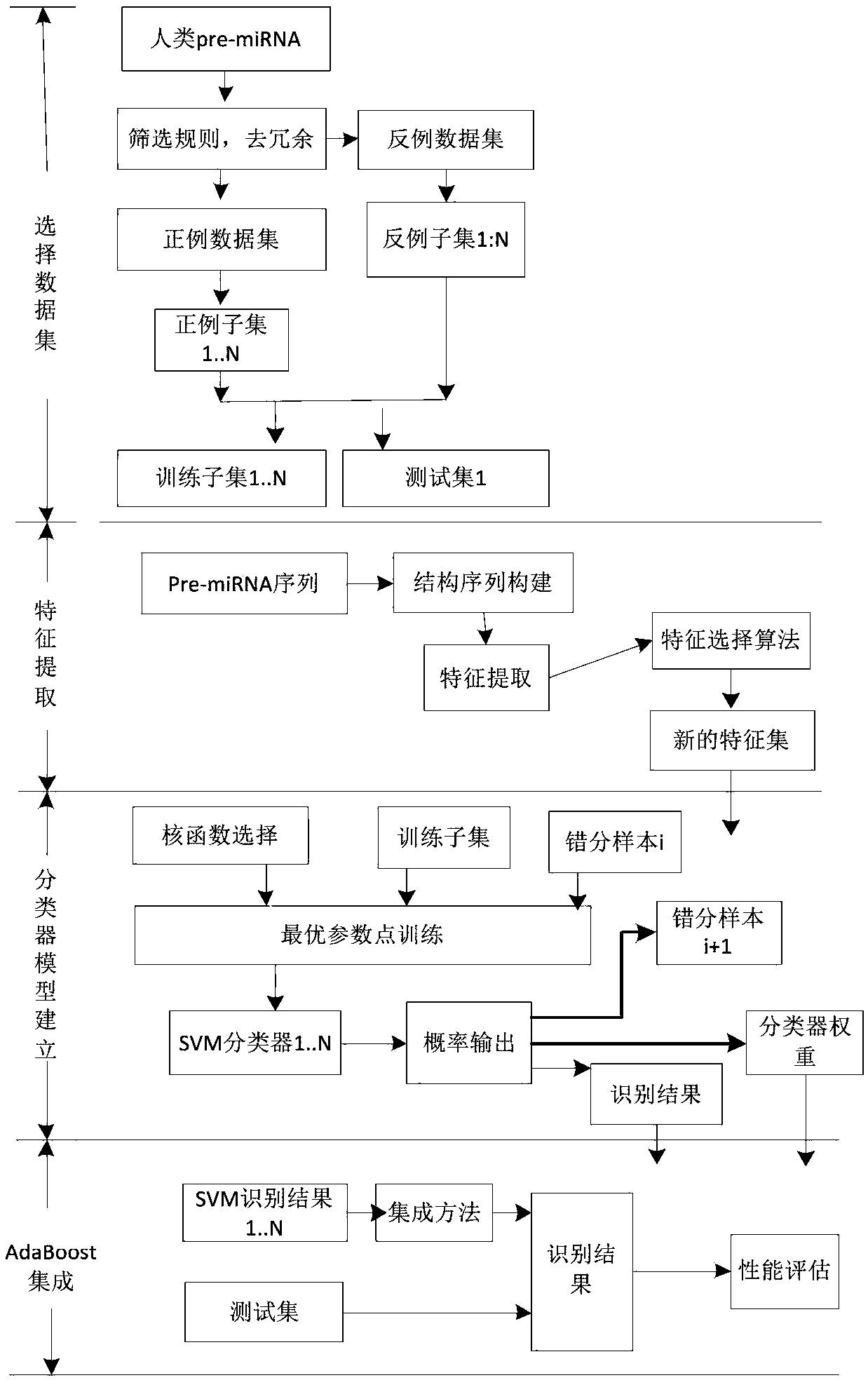
[0027] The mature miRNA all-site recognition method based on SVM-AdaBoost of the present embodiment, described recognition method is realized through the following steps:
[0028] Step 1, select the pre-miRNA sequence in the miRBase database, and set up a training data set and a test set on the selected sequence;
[0029] Step 2. Extract the biological characteristics of mature miRNA splicing sites based on the structured sequence:
[0030] Step 21. Based on the biometric analysis, define the biometric characteristics of the mature miRNA cleavage site;
[0031] Step 22, defining the mature miRNA duplex and the site corresponding to the mature miRNA duplex;
[0032] Step two and three, constructing a sequence on the defined mature miRNA duplex for feature extraction;
[0033] Step 24, predicting the secondary structure and free energy of the constructed sequence;
[0034] Step 25, extracting a feature set on the constructed sequence;
[0035] Step 3. Obtain a new feature se...
specific Embodiment approach 2
[0039] Different from the specific embodiment one, in the mature miRNA all-site recognition method based on SVM-AdaBoost of the present embodiment, the pre-miRNA sequence in the miRBase database is selected in step one, and a training data set is established on the selected sequence and the procedure for the test set is,
[0040] Select the pre-miRNA sequence in the miRBase database, remove redundant sequences and multi-branched sequences, and establish a training set and a test set for the 3' end and a training set and a test set for the 5' end in the remaining sequences; among them, the pre-miRNA The meaning is precursor miRNA;
specific Embodiment approach 3
[0041] The difference from Embodiment 1 or Embodiment 2 is that in the SVM-AdaBoost-based full-site recognition method of mature miRNA in this embodiment, the pre-miRNA sequence in the miRBase database is selected in step 1, which is a human pre-miRNA sequence.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com