SNP selection method based on improved fuzzy clustering algorithm
A fuzzy clustering and algorithm technology, applied in the field of SNP selection, can solve the problems of no further mining of associations, research troubles, no differences, etc., and achieve the effect of excellent classification effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
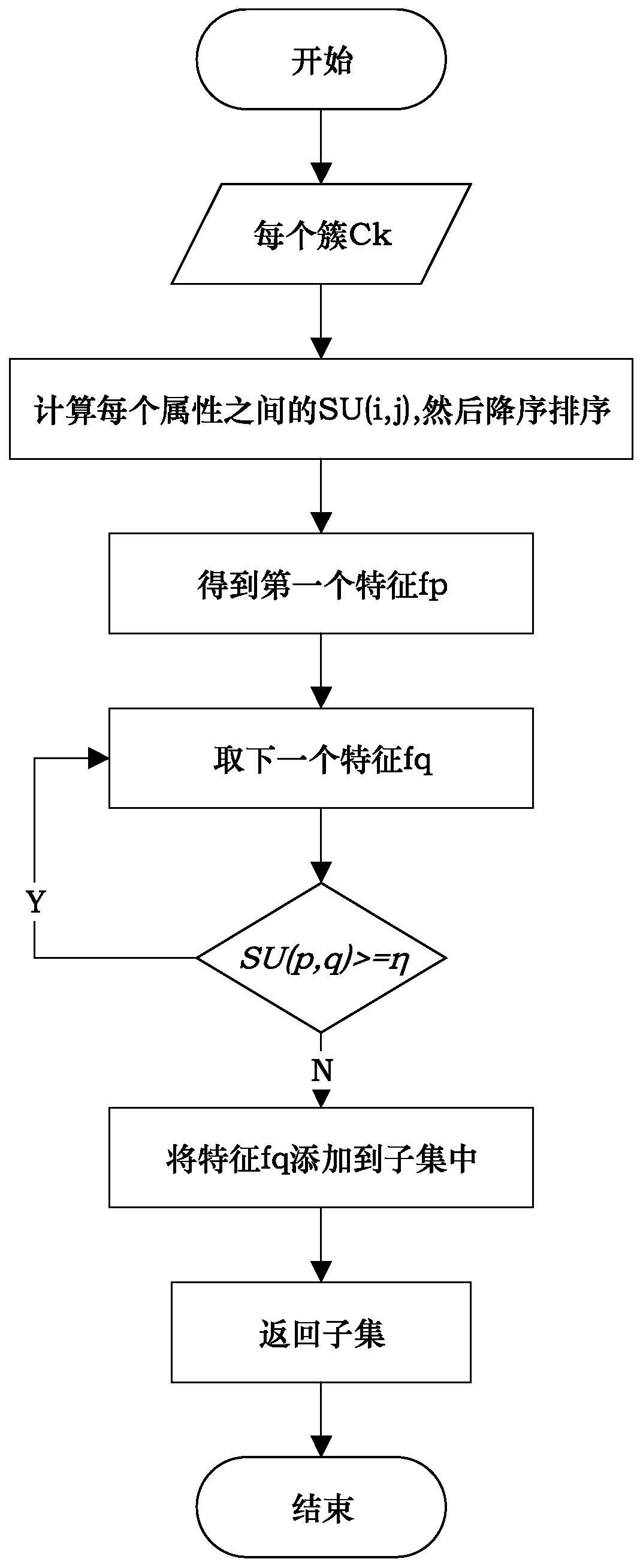
[0054] A SNP selection method based on an improved fuzzy clustering algorithm. For SNP data, while considering the impact of a single SNP on the classification results, it also takes into account the interrelationships between SNPs in local areas, and realizes dimensionality reduction for data. At the same time, the information inside the SNP was fully excavated. Specifically include the following steps:
[0055] Step 1. Obtain the SNP data set. Generally, the original data is expressed in the form of genotype, such as AT, GC, AA...CG, etc.
[0056] Step 2, preprocessing the SNP data to obtain the preprocessed data, the preprocessing mainly includes the processing of missing values and data recoding; the details are as follows:
[0057] 1): First of all, for each SNP, the absence of genotype representation can be counted. If the missing ratio is higher than the set threshold (here set to 20%), the corresponding SNP will be deleted from the data set.
[0058] 2): For the de...
Embodiment 2
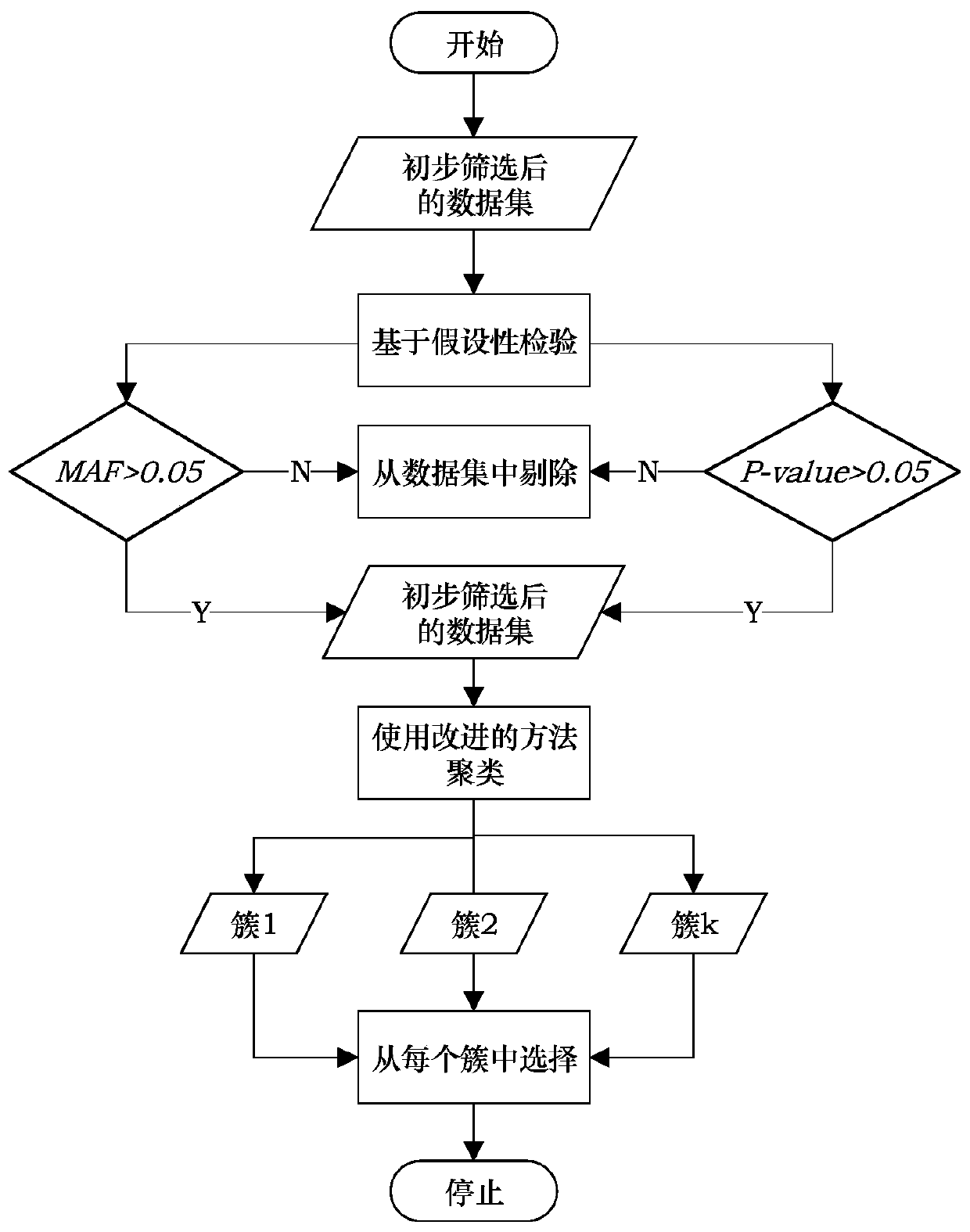
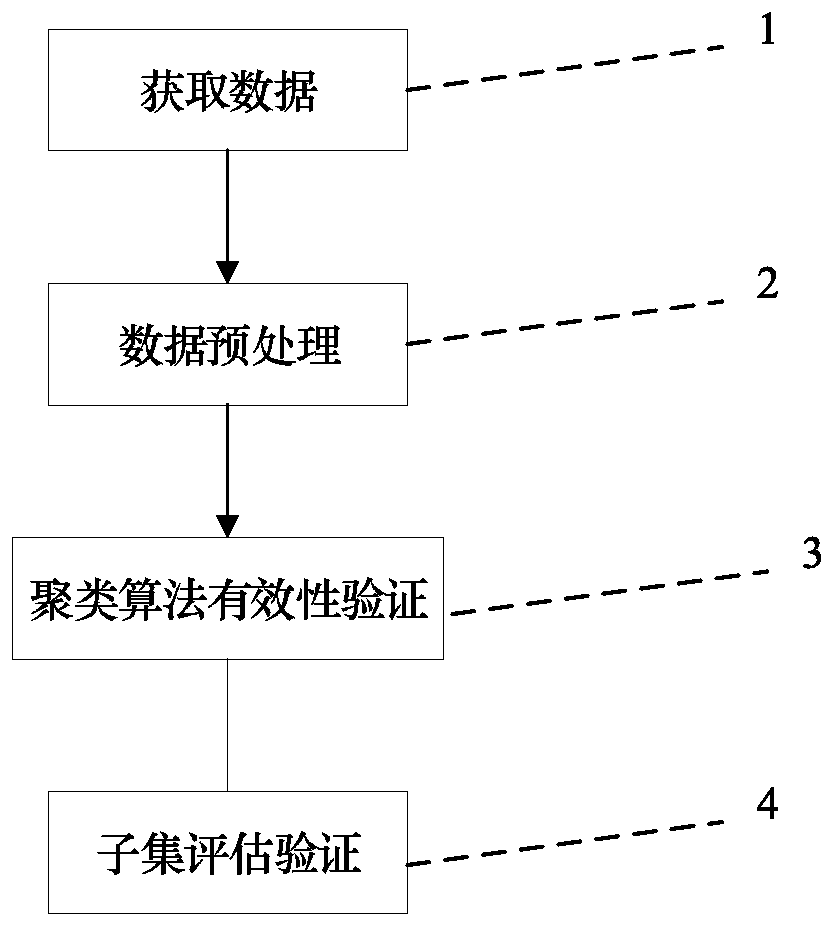
[0102] It is verified by experiments that the SNP subset constructed by this method has a better classification effect than other selection methods, and can be applied to the selection of SNP data. Use clinical data to verify (select part of the data, and the data is recorded as G1000), the experiment is implemented as follows image 3 As shown, it specifically includes the following parts:
[0103] The data preprocessing unit 2 is used for preliminary screening of data based on hypothesis testing. The threshold of MAF is set to 0.05, and the results show that the MAF values of the dataset G1000 are all greater than 0, so there is no need to delete any SNP; the threshold of the p-value of the chi-square test is set to 0.03, and the results show that there are 228 SNPs that do not meet this condition. to delete.
[0104]The clustering algorithm effectiveness evaluation verification unit 3 is used to evaluate the clustering method proposed by the present invention, specifica...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com