Prediction method of a G-Protein Coupled Receptor (GPCR) drug and a targeting pathway based on heterogeneous network
A prediction method and heterogeneous network technology, applied to the analysis of two-dimensional or three-dimensional molecular structure, biostatistics, bioinformatics, etc., can solve problems such as no direct prediction of DPI
Active Publication Date: 2019-07-16
EAST CHINA NORMAL UNIV
View PDF5 Cites 8 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
The disadvantage of the method is that changing the sign of the impact factor at the same time will not cause the expected impact on the estimated gene expression value, and the data provided are the sensitivity data of the drug and the gene expression data in the cell line cultured by the drug ( References Ma H, Zhao H.iFad:an integrativefactor analysis model for drug-pathway association inference[J].Bioinformatics,2012,28(14):1911-8.MaH,Zhao H
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
 Login to View More
Login to View More PUM
 Login to View More
Login to View More Abstract
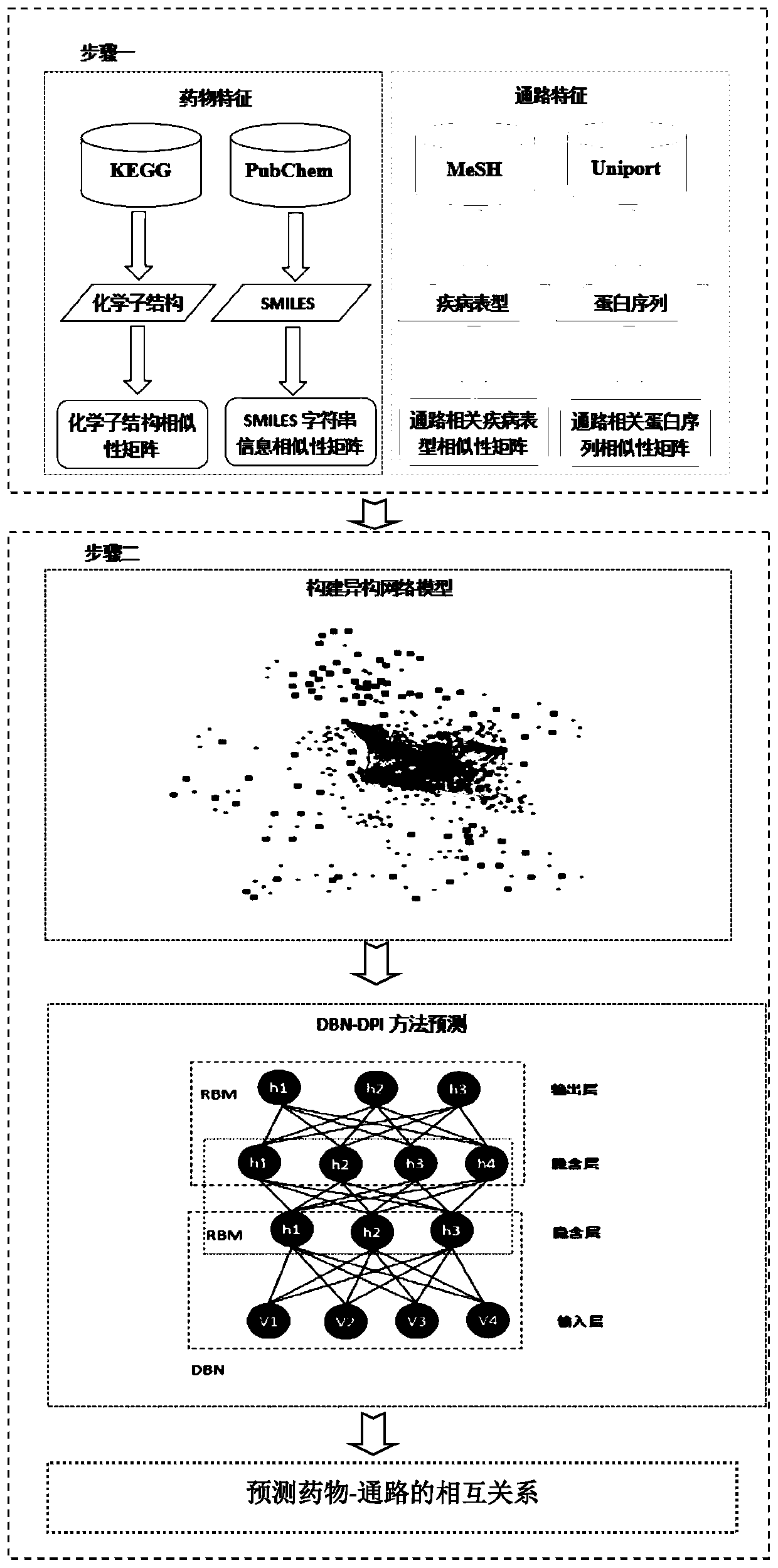
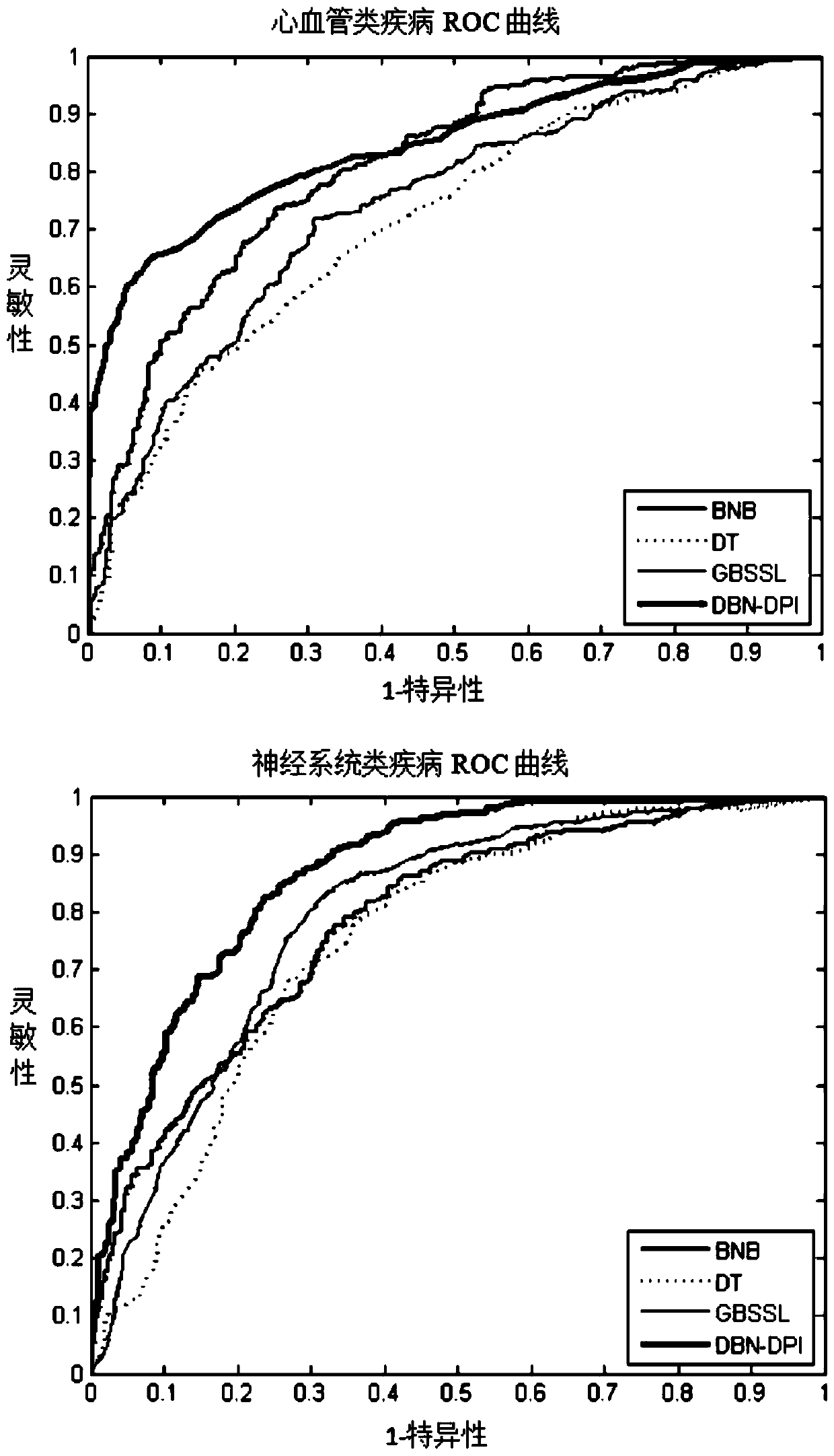
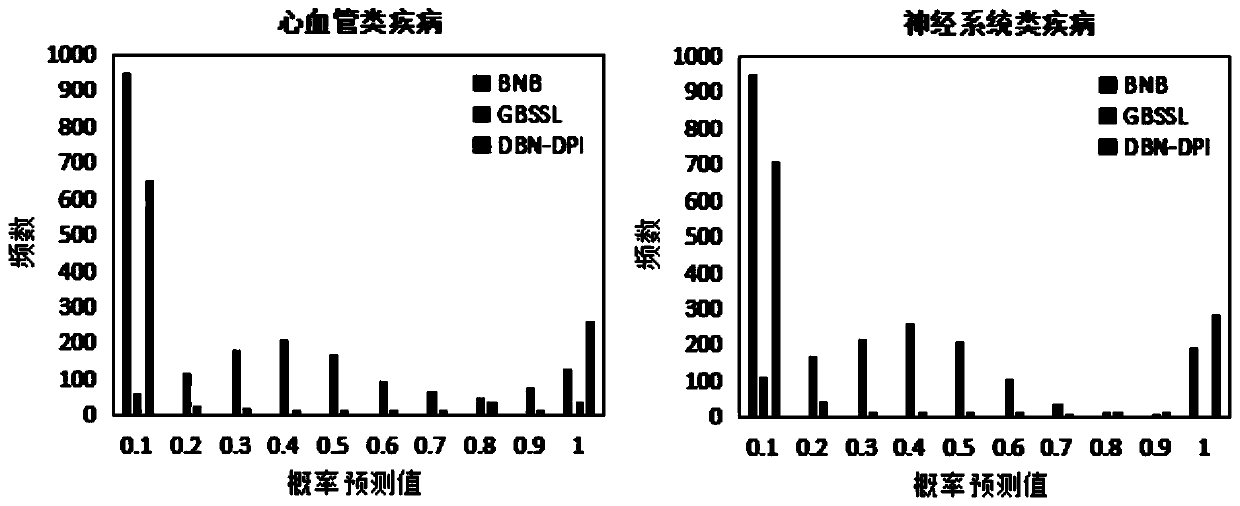
The invention discloses a prediction method of a G-Protein Coupled Receptor (GPCR) drug and a targeting pathway based on a heterogeneous network and biological application of the method. The method ischaracterized in that a drug-pathway heterogeneous network model is built based on the chemical sub-structure feature of the drug, the SMILES character string information feature of the drug, pathway-related protein sequence features and the phenotypic features of pathway-related diseases, and a drug-pathway interaction (DPI) relationship is predicted by using a deep belief network method. The prediction method of the GPCR drug and the targeting pathway based on the heterogeneous network, provided by the invention, not only is high in prediction precision and robustness, but also can successfully predict the pathway which part of the GPCR drug acts on. The method can be applied to drug-pathway correlation evaluation, clinical patient medication reference and the like.
Description
technical field [0001] The invention relates to the connection between drugs and pathways in bioinformatics, that is, a method for predicting GPCR drugs and targeted pathways based on a heterogeneous network model. The method mainly uses the chemical substructure characteristics of drugs and the SMILES string information characteristics of drugs, And pathway-related disease phenotype similarity and pathway-related protein sequence similarity, so as to construct a drug-pathway heterogeneous network model, and use the deep belief network method to predict potential drug-pathway relationships. Background technique [0002] Identifying drug-pathway interactions (DPI) is the key to drug discovery and drug repositioning. Due to the clear availability of GPCR drugs, if the drug can be used in a new pathway, it can not only reduce the cost of drug development, but also reduce the adverse reactions of the drug. Although various bioassay techniques are currently available to predict ...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More IPC IPC(8): G16B15/30G16B40/00G16B50/00
CPCG16B15/30G16B40/00G16B50/00Y02A90/10
Inventor 江振然蒋惠炎
Owner EAST CHINA NORMAL UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com