Method for inserting turnip mosaic virus (TuMV) P3 protein into labels and recombinant vector and application of (TuMV) P3 protein
A technology of turnip mosaic virus and recombinant vector, applied in the field of genetic engineering, can solve the problems of not necessarily coincident distribution of expression levels, complex interaction of virus proteins, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
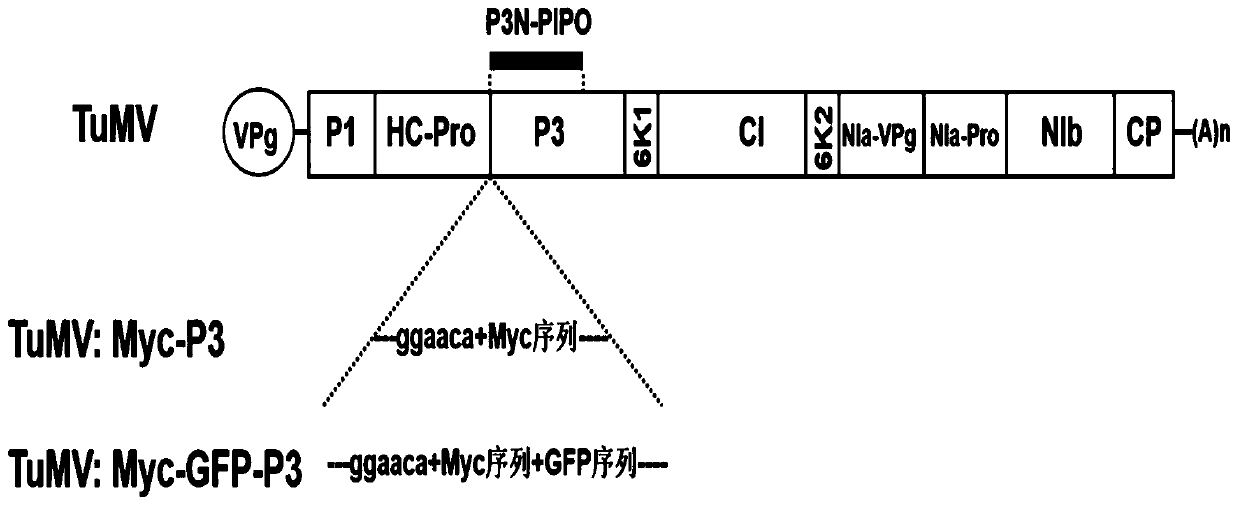
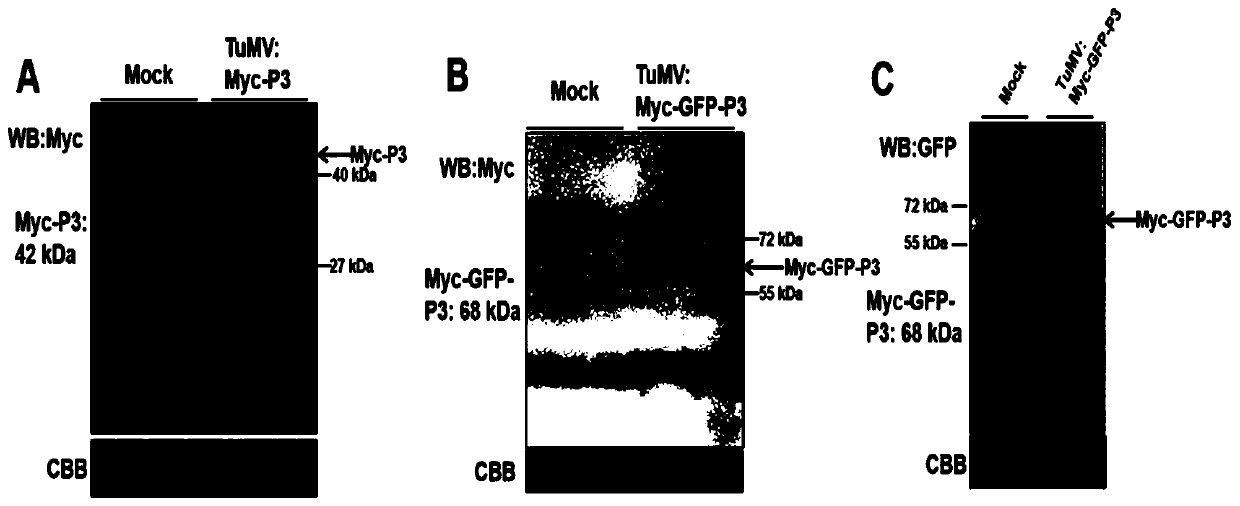
[0052] Such as figure 1 Shown is a method for inserting tags into the P3 protein of TUMOV, inserting Myc into the N-terminus of pCambia2300-TuMV P3 protein. Specifically include the following steps:
[0053] (1) Use TuMV-KpnI-F and Hcpro-2a-Myc-P3-R as primers, and use the plasmid containing the TuMV sequence as a template to amplify the first specific PCR band by PCR with high-fidelity DNA polymerase , and recover a specific PCR band, called PCR1;
[0054] TuMV-KpnI-F: tacgagacaagagc ggtacc aaatgggtcacggaaac,
[0055] TuMV-SnaBI-R: aagacatactac tacgta atctattacctatac, as shown in SEQ ID: No.3-4;
[0056] (2) Using Hcpro-2A-Myc-P3-F and TuMV-SnaBI-R as primers, and using the plasmid containing the TuMV sequence as a template, high-fidelity DNA polymerase was used to amplify the second specific PCR band by PCR , and recover a specific PCR band, called PCR2;
[0057] Hcpro-2A-Myc-P3-F:acactaccgcgttggaggaacagagcagaagctgatctcagaggaggacctgggaacagaatgggaggac,
[0058] Hcpr...
Embodiment 2
[0065] Such as figure 1 Shown is a method for inserting tags into the P3 protein of TUMOV, inserting Myc-GFP into the N-terminus of the pCambia2300-TuMV P3 protein. Specifically include the following steps:
[0066] (1) Using TuMV-KpnI-F and GFP-Myc-Hc-R as primers, using a plasmid containing the TuMV sequence as a template, using a high-fidelity DNA polymerase to perform PCR amplification to obtain the first specific PCR band, and Recover a specific PCR band, called PCR1;
[0067] TuMV-KpnI-F: tacgagacaagagc ggtacc aaatgggtcacggaaac,
[0068] GFP-Myc-Hc-R: aagttcttctcctttactcaggtcctcctctgagatcagcttctgctctgttcctccaacgcggta, as shown in SEQ ID: No.3 and No.8;
[0069] (2) Use Hc-Myc-GFP-F and P3-2A-GFP-R as primers, and use the plasmid containing the GFP sequence as the template, and use high-fidelity DNA polymerase to perform PCR to amplify the second specific PCR band , and recover a specific PCR band, called PCR2;
[0070] Hc-Myc-GFP-F: taccgcgttggaggaacagagcagaagctga...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com