Zinc enzyme docking method based on optimal geometric matching
A geometric and optimal technology, applied to the analysis of two-dimensional or three-dimensional molecular structure, bioinformatics, informatics, etc., can solve the problem of inability to accurately predict the docking position of small drug molecules and zinc enzymes, and the inability to obtain small molecules Docking results with zincase and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
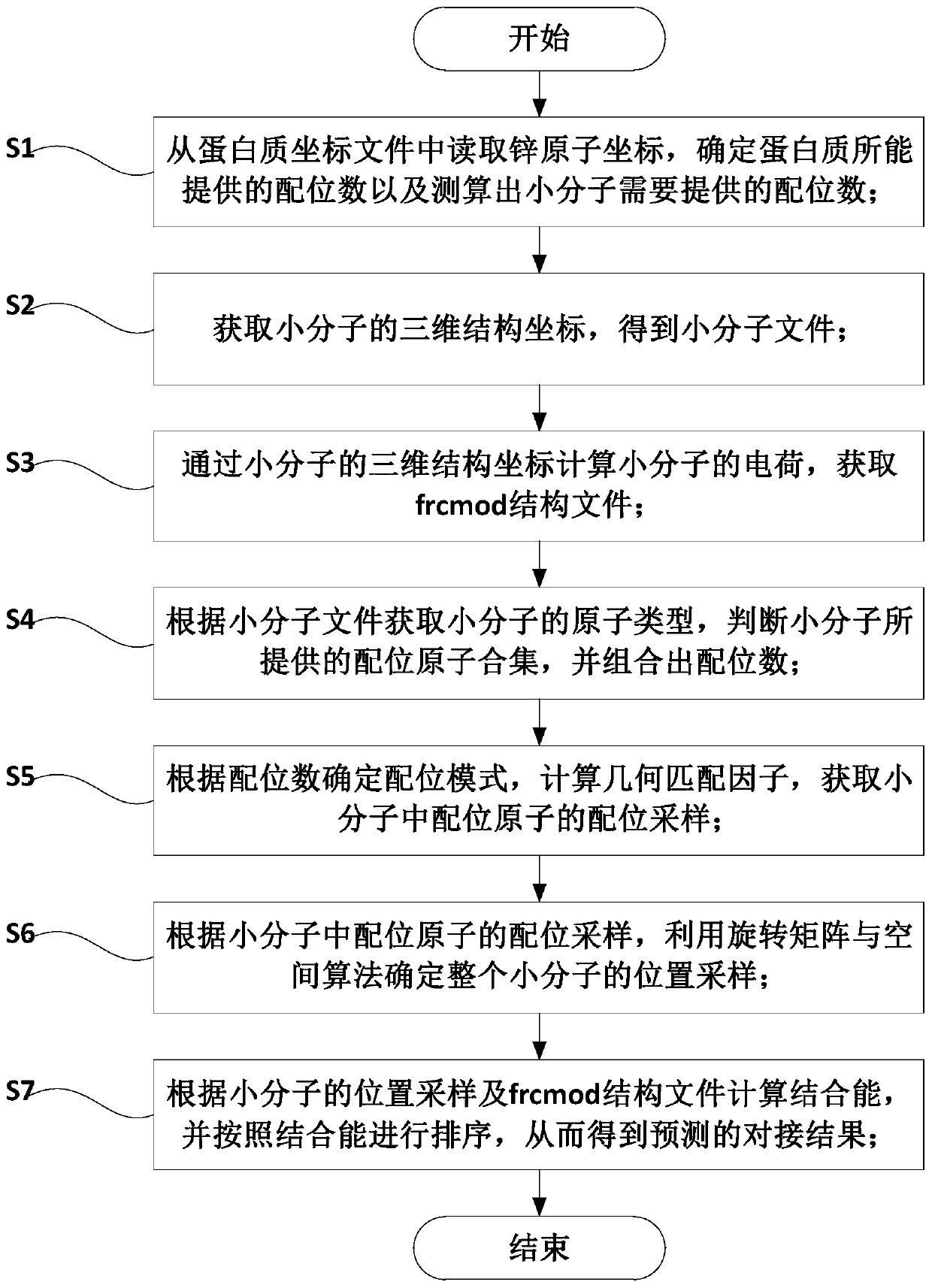
[0047] like figure 1 Shown, a zinc enzyme docking method based on optimal geometric matching, including the following steps:
[0048] S1: Read the zinc atom coordinates from the protein coordinate file, determine the coordination number that the protein can provide and calculate the coordination number that the small molecule needs to provide;
[0049] S2: Obtain the three-dimensional structure coordinates of the small molecule, and obtain the small molecule file;
[0050] S3: Calculate the charge of the small molecule through the three-dimensional structure coordinates of the small molecule, and obtain the frcmod structure file;
[0051] S4: Obtain the atom type of the small molecule according to the small molecule file, judge the set of coordination atoms provided by the small molecule, and combine the coordination numbers;
[0052] S5: Determine the coordination mode according to the coordination number, calculate the geometric matching factor, and obtain the coordination...
Embodiment 2
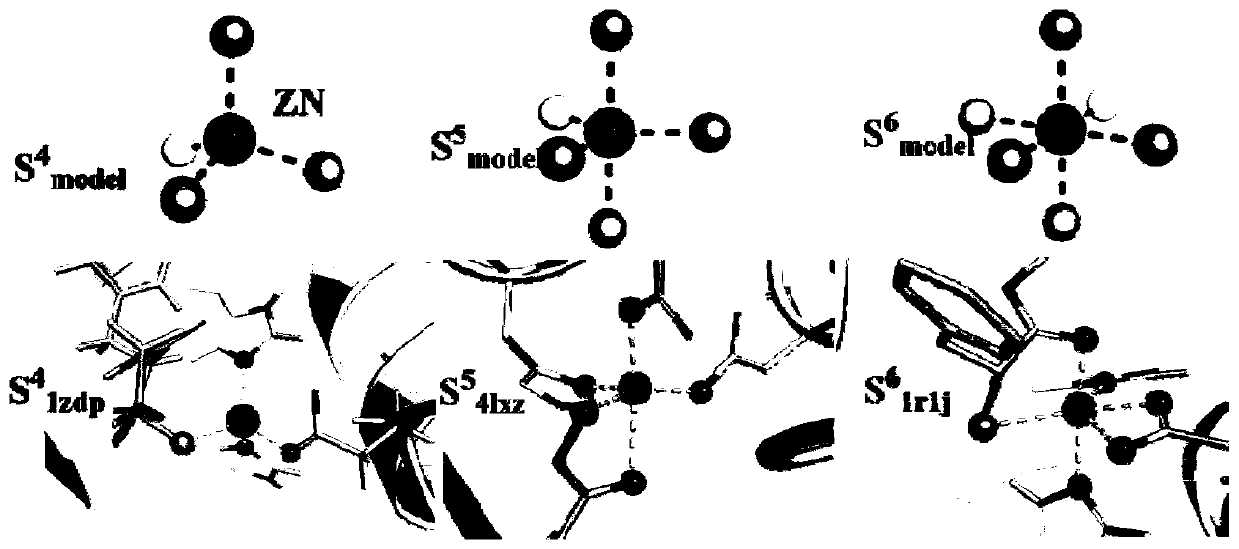
[0080] On the basis of Example 1, redock experiments were carried out on 53 zinc proteins. Under the three conditions of ensuring the coordination number, the same coordination atoms and RMSD That is, the correct docking results of the Glide XP algorithm and Autodock4 are 14 and 13 respectively, while the correct docking results of the present invention are 45, and the accuracy is much higher than that of traditional docking software.
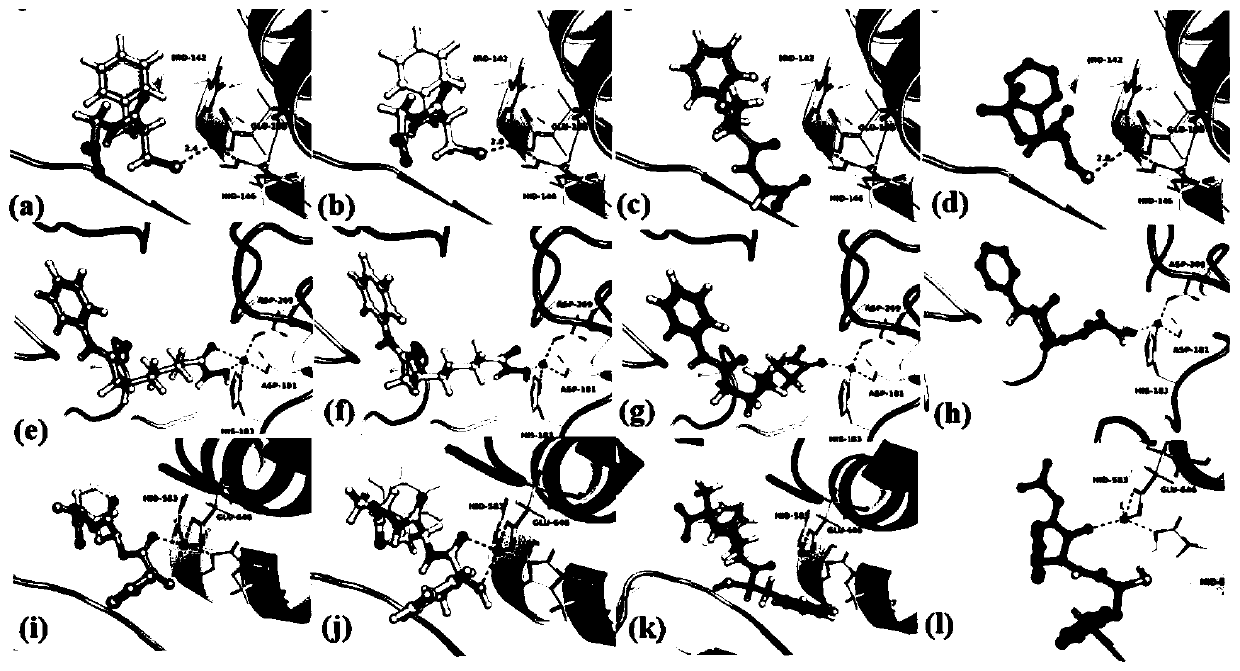
[0081] In the specific implementation process, such as image 3 As shown, Fig. a, Fig. e and Fig. i are three crystal structures, respectively corresponding to four-coordination 1zdp, five-coordination 4lxz and six-coordination 1r1j; Fig. b, Fig. f and Fig. The docking prediction results of the corresponding proteins with five and six coordinations, the coordination number and the coordination distance prediction effect are better; Figure c, Figure g and Figure k are Even using the Glide XP algorithm for the docking prediction results of the c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com