SNP locus related to wheat spikelet number per spike and application thereof
A spikelet number and locus technology, applied in the field of SNP loci, can solve the problems of small contribution rate of phenotype, poor repeatability, difficulty in applying wheat genetic improvement, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
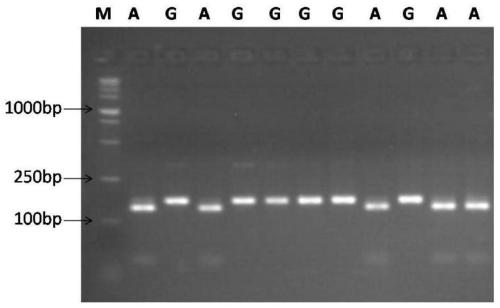
[0026] Example 1 SNP and its PCR-enzyme polymorphism detection related to the number of spikelets per ear of wheat
[0027] 1. Specific primers and sequence analysis for amplifying the genomic fragment containing the SNP in wheat
[0028] A SNP was found in the gene WFZP promoter region on the wheat genome, corresponding to the 1501st position from the 5' end of Sequence Table 1; there are two genotypes at this site in the wheat natural variation population:
[0029] Genotype A: A
[0030] Genotype B: G
[0031] According to the sequence differences of different genomes of wheat, specific primers were designed to PCR amplify the DNA fragments respectively including the SNP site:
[0032] F1: GCAGGCATCTTTTACACCATCTTA
[0033] R1: GGCAGAAGTGAAGTGAGGT
[0034] F2: GCAAGGATTGTGGCATGCA
[0035] R2: CAAAGAGATGAGGAGAATGTTGAG
[0036] Using F1 and R1 as primers for the target sequence of PCR amplification as shown in the sequence sequence 1 of the sequence listing 1083-1887 sequ...
Embodiment 2
[0059] In 2015, the above-mentioned natural group wheat was planted in the dry land of the Experimental Farm of the Institute of Crop Science of the Chinese Academy of Agricultural Sciences (Shunyi, Beijing), and in 2016 in the dry and wet land of the Experimental Farm of the Institute of Crop Science of the Chinese Academy of Agricultural Sciences (Shunyi and Changping, Beijing). The number of spikelets per ear of each wheat variety is associated with the situation of the number of spikelets per ear and the polymorphic site with Tassel2.1 software, and the mixed linear model+population structure (MLM+(Q+K)) is selected Methods for analysis, with P<0.05 as the significance level, the results are shown in Table 2.
[0060] Table 2 Association analysis results of WFZP-A gene polymorphisms and number of spikelets per panicle in natural populations
[0061]
[0062]
[0063] The results of the association analysis in Table 2 showed that the difference in the number of spikel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com