Metagenome characteristic selecting method based on variable importance scoring and Neyman-Pearson detection
A feature selection method, metagenomic technology, applied in the field of metagenomic feature selection based on variable importance score and Neiman Pearson test, can solve the problem that random forest is susceptible to noise, achieve good stability and classification effect, Good robustness, convenient medical verification test effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
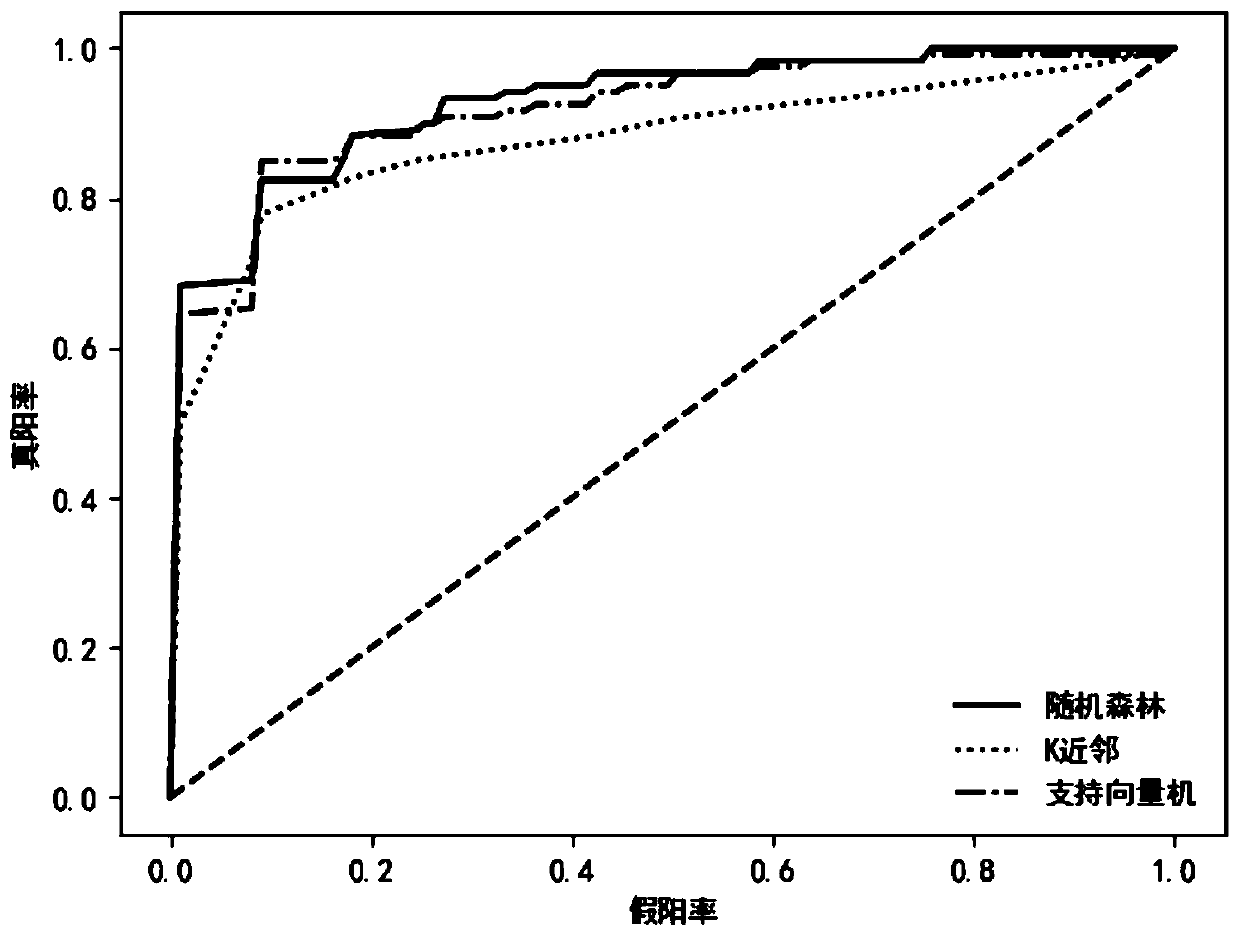
[0025] The present invention will be further described below in conjunction with the accompanying drawings and embodiments. This embodiment is aimed at the cirrhosis of the liver (Cirrosis of Liver, CIR) metagenomic data set. The cirrhosis data set is collected from the intestinal tract, including 232 samples, including 118 cases of cirrhosis. There were 114 cases in the control group, involving 532 operable taxa.
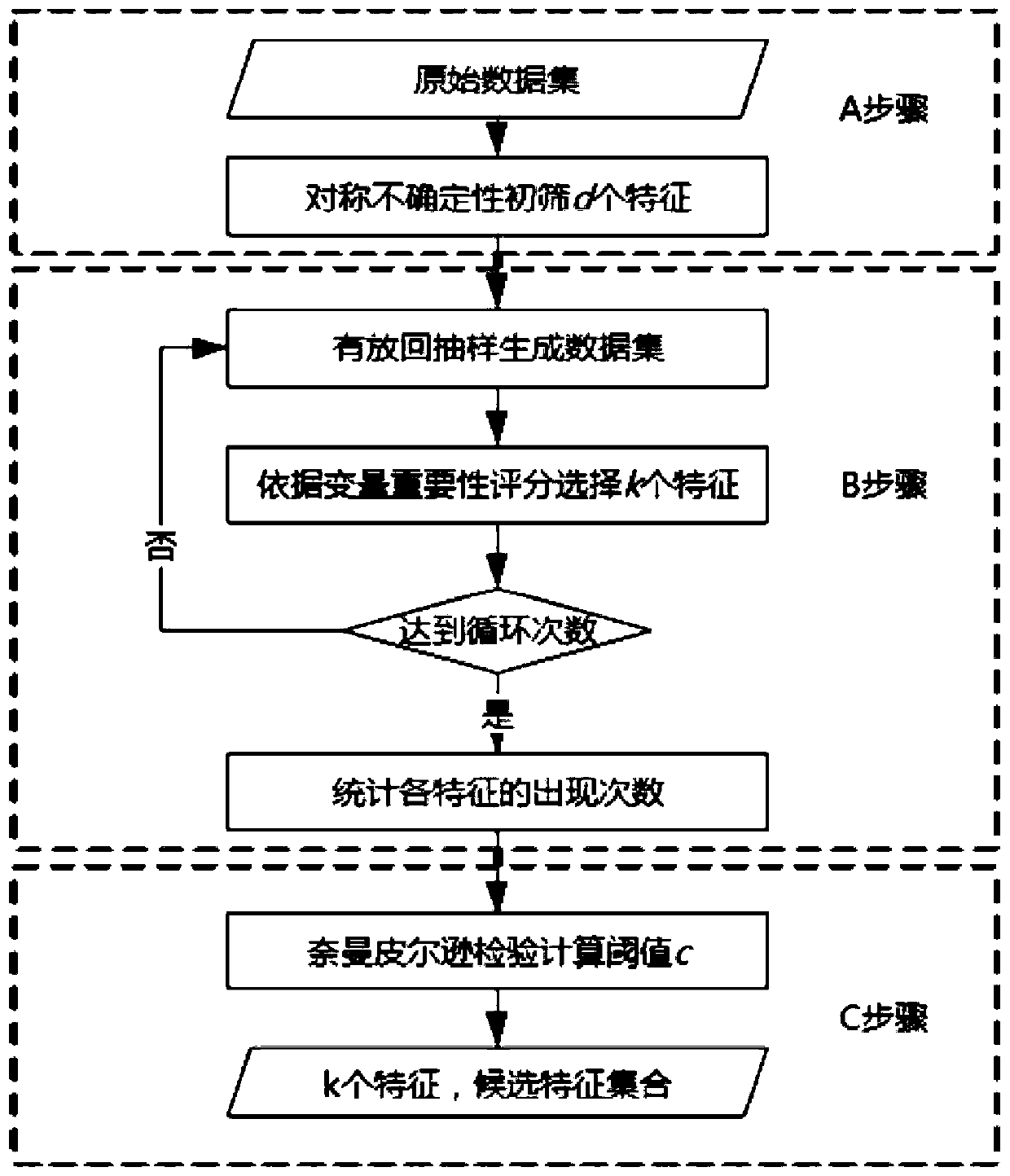
[0026] refer to figure 1 , a metagenome feature selection method based on variable importance scores and Neyman-Pearson tests, including the following steps:
[0027] Step A: For the cirrhosis operational unit dataset, calculate the correlation of each microbial signature with the sample phenotype using symmetric uncertainty, sort by size, select the top 200 features as a feature subset, and generate subdata of the original data set for subsequent analysis.
[0028] Step B: First sample the sub-dataset with replacement sampling, then calculate the variable import...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com