Giant baby forecasting model based on peripheral blood free DNA detection
A prediction model and peripheral blood technology, applied in the field of macrosomia prediction model, can solve problems such as inaccurate prediction of macrosomia, and achieve good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Model method for predicting macrosomia based on peripheral blood cell-free DNA
[0046] The method of the present invention for predicting macrosomia based on free DNA in peripheral blood is as follows: compare the sequencing results of free DNA in peripheral blood with the genome sequence map, and then calculate the number of DNA fragments from the transcription start site region of the gene to be tested in the same sample , corrected according to the total number of DNA sequences, and after normalizing the abundance of free DNA, use machine learning algorithms to calculate and output the prediction results of macrosomia in pregnant women through the optimal combination of different differential genes, which can effectively predict macrosomia onset.
[0047] Specifically, the method steps are as follows:
[0048] Step 1: Determine where the DNA fragments in the plasma come from on the chromosomes
[0049] A control study was conducted with pre-morbid sample...
Embodiment 2
[0080] Embodiment 2 sample detection example
[0081] 1. Experimental sample:
[0082] The training group included 119 macrosomia samples and 378 healthy controls;
[0083] The validation group included 72 samples of macrosomia and 162 healthy controls.
[0084] According to the method operation of embodiment 1. Accuracy, sensitivity and specificity of statistical calculation methods.
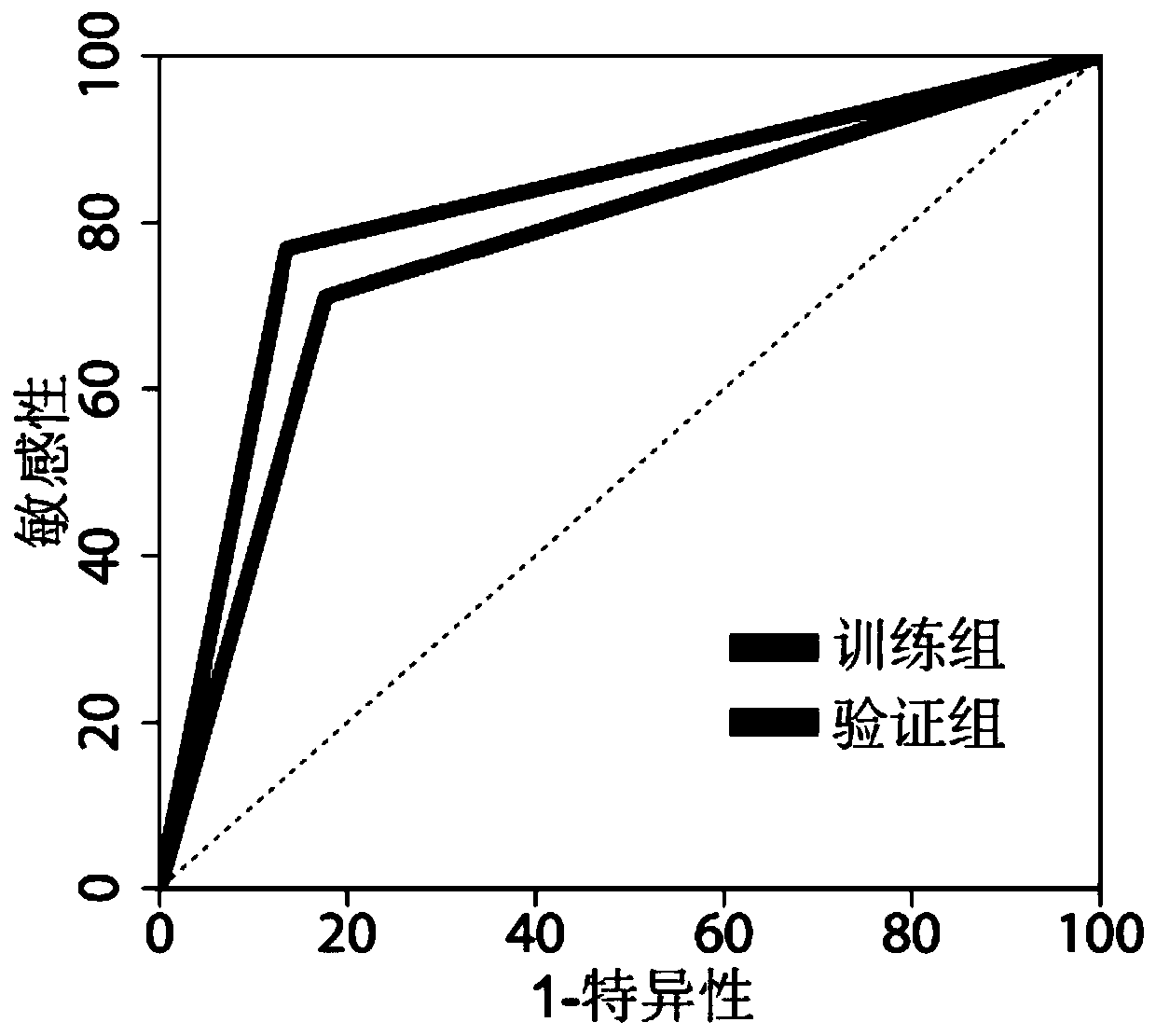
[0085] 2. The results show that the method model of the present invention can effectively judge macrosomia pregnant women before the early onset in the training group and the verification group (table 4 and figure 2 ).
[0086] Table 4
[0087]
[0088] Among them, the calculation result example is as follows:
[0089] Sample 1 (pre-onset sample of diagnosed macrosomia):
[0090] logit(Y)=2.180+0.605×SMC3–1.204×MASTL+1.366×CREM–1.295×C1QTNF12–0.471×MLXIP–0.811×MAP3K9–1.284×IGSF6–1.347×APC2–0.504×GPM6A+1.048×TMEM128×NIP.057 –1.652×TMEM184A=2.755
[0091] Y=0.940
[0092] If the sam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com