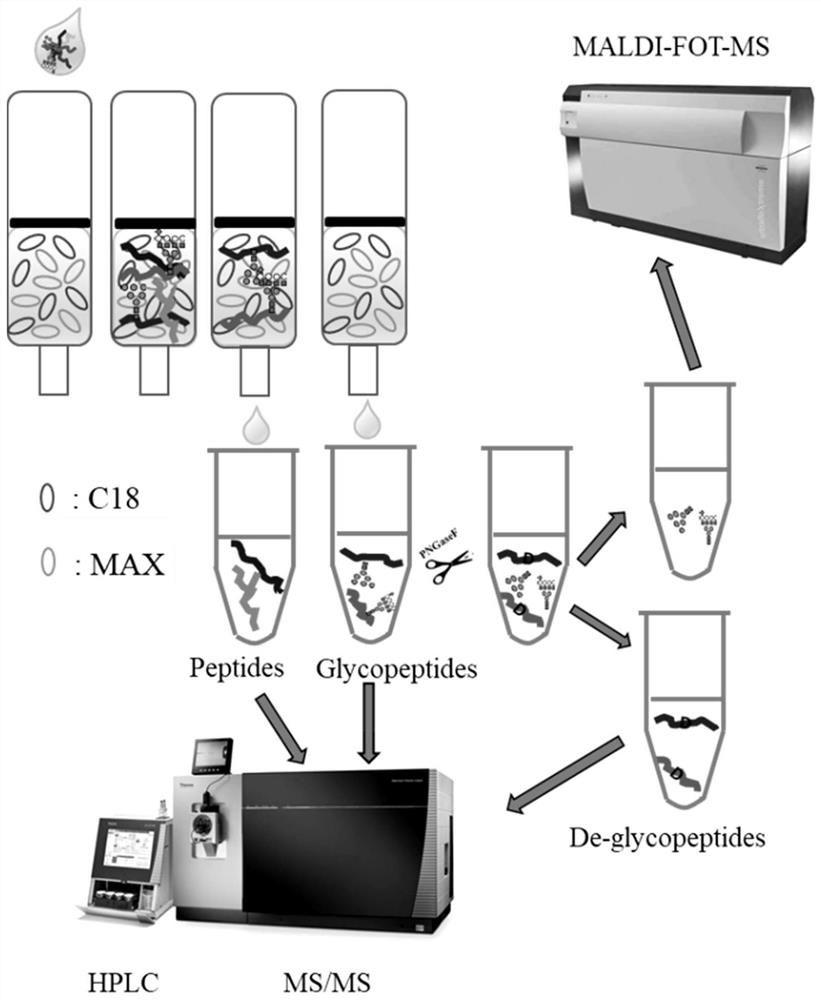
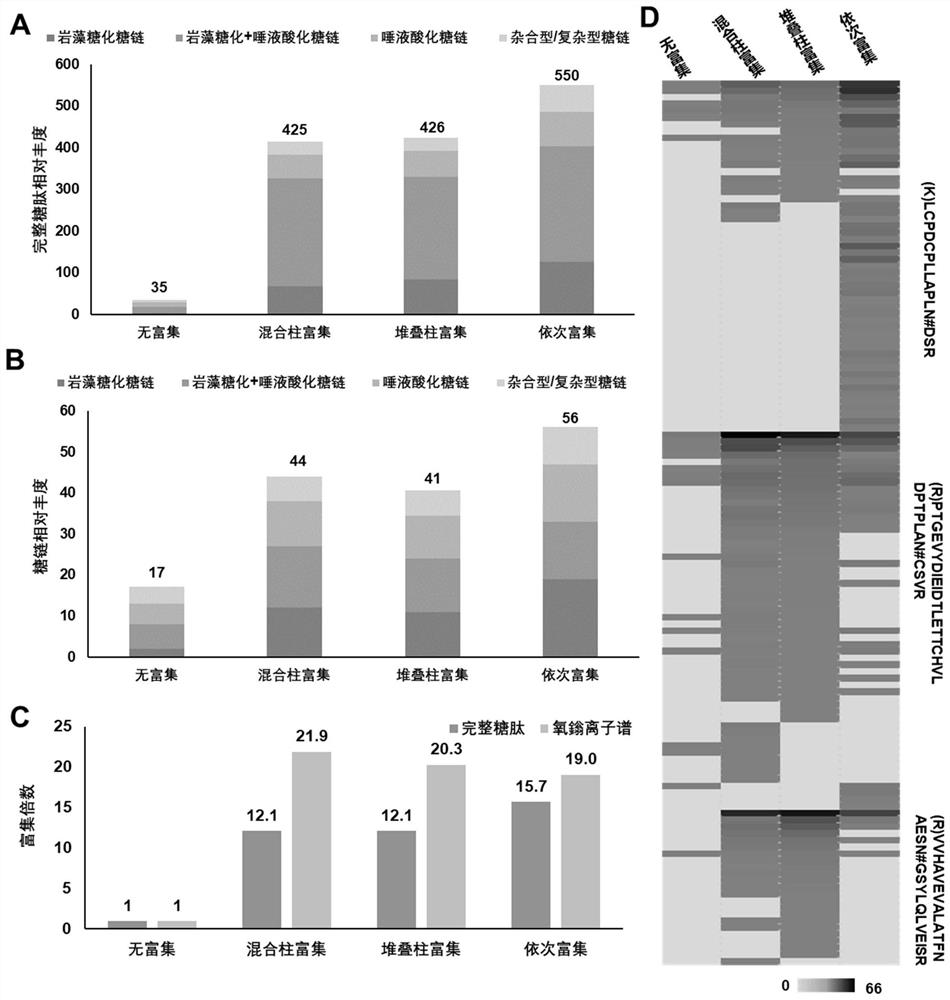
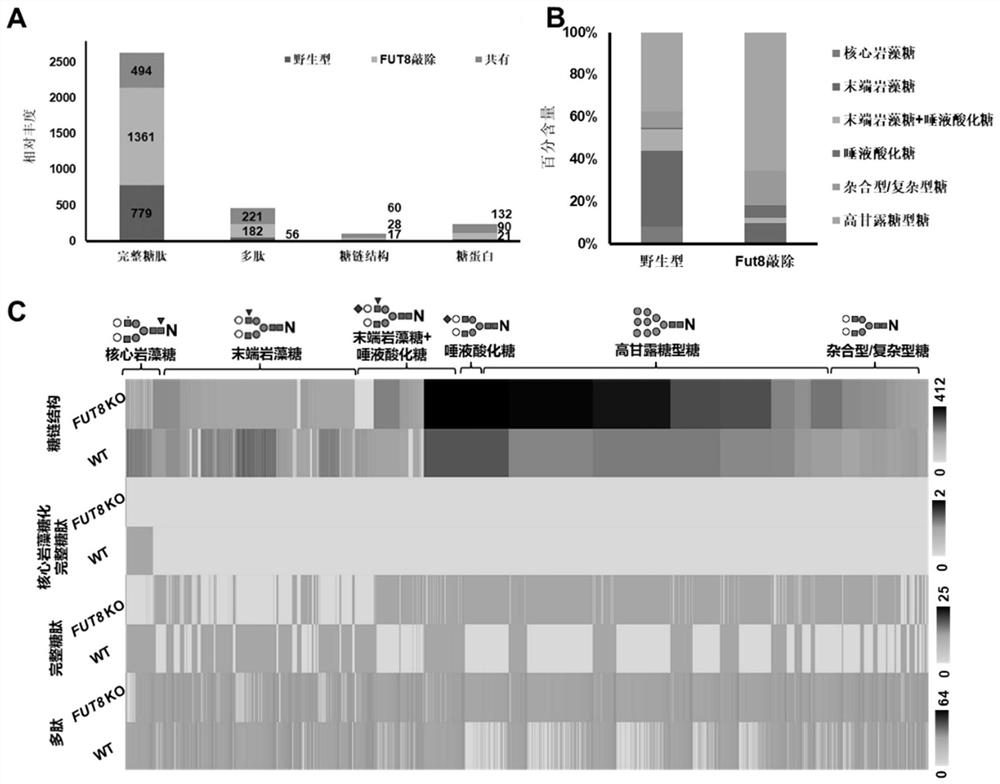
A rapid method for analyzing proteins and highly polar long amino acid sequence glycopeptides
A rapid analysis, protein technology, applied in the field of glycopeptide analysis, can solve the problems of complete glycopeptide analysis efficiency and coverage reduction, sample loss, long time consumption, etc., and achieve the effect of high enrichment and identification efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Sample protein preparation:
[0033] Preparation of cell / tissue protein: Use a cell protein reagent extraction kit (such as T-PER, RIPA) to extract cell protein, use as little lysate as possible to ensure a protein concentration of 2-10 mg / mL, and centrifuge at 14,000g for 5 minutes to remove cell debris . The protein concentrations of the two cell extracts were determined with a BCA kit (guaranteed at least three repetitions).
[0034] Serum protein preparation: Serum samples were centrifuged at 12,000g for 15 minutes, and the middle liquid part was taken. Add to a 10KD ultrafiltration tube, place the ultrafiltration tube in a matching centrifuge tube, centrifuge at 12,000g for 15min, add 400μl of 40mM NH 4 HCO 3, centrifuge again, and repeat. Invert the filter membrane into a new centrifuge tube, centrifuge at 9000g for 3min, collect the separated protein (about 50μl), quantify it by Bradford method, and finally adjust the volume to 2mg / ml respectively.
[0035] ...
Embodiment 2
[0056]Analysis of WT and Fut8 knockout (Fut8 - / - ) Complete glycopeptides of glycoengineered CHO cells:
[0057] We applied the one-step enrichment method of Example 1 to the analysis of wild-type (WT) and glycoengineered Chinese hamster ovary (CHO) cells, and evaluated the N-linked glycosylation and even Changes in glycoprotein expression. By knocking out the Fut8 gene in CHO cells (Fut8 - / - ) can generate core fucosylation-deficient cell lines. Fut8 (α1,6-fucosyltransferase) catalyzes the transfer of a fucose residue to position 6 of the innermost GlcNAc residue of an N-linked oligosaccharide of a glycoprotein (ie, core fucosylation). Based on the glycopeptides obtained by the one-step enrichment method, after LC-MS / MS analysis, the screening criteria for the database searched by GPQuest are as follows: (1) glycopeptide error rate (FDR) is less than 1%, (2) each peptide requires PSM ≥2, (3) all PSMs should annotate at least one N-linked glycan, (4) the core Fuc fragment ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap