RT-PCR detection method and kit for novel coronavirus
A RT-PCR, coronavirus technology, applied in the field of biotechnology and molecular diagnosis, can solve the problems of unfavorable virus discovery and prevention, mutant virus missed detection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] The design of embodiment 1 primer and probe
[0093] Based on the sequence of the novel coronavirus, the conserved regions of its genome were analyzed. Dozens of specific primers and probe sequences for the detection of the 2019-nCoV E gene were designed in these conserved regions. In addition, in order to monitor the process of specimen collection, nucleic acid extraction and PCR amplification, ribonuclease P (RPP30) in human genomic DNA was selected to design internal standard primers and probes.
[0094] During the design process of the above primers and probes, try to avoid the formation of hairpin structures, internal dimers of primers, dimers between primers and mismatches.
[0095] In addition, by comparing the NCBI Blast online database (https: / / blast.ncbi.nlm.nih.gov / Blast.cgi) to the sequence of the above-designed novel coronavirus-specific primers and probes to avoid comparison with other viruses or Non-specific tuberculosis occurs in human genes. Through ...
Embodiment 2
[0099] Embodiment 2 Inspection method and system
[0100] The principle of the Taqman probe used to detect the new coronavirus RNA is: first use the added primer sequences E-NCOV-F and E-NCOV-R as reverse transcription primers to reverse-transcribe the extracted viral RNA into The cDNA was then amplified using E-NCOV-F and E-NCOV-R as forward and reverse primers for real-time fluorescent PCR detection.
[0101] This method is a one-step amplification method, which combines two steps of reverse transcription of viral RNA into cDNA and DNA amplification, and does not need to change tubes for operation.
[0102] In order to determine the detection system of primers and probes, the effects of different concentrations of primers and probes on the fluorescent PCR reaction were tried. The result shows that when E-NCOV-F, the concentration of E-NCOV-R primer is 10pmol, the concentration of E-NCOV-P probe is 5pmol, RP30-F, the concentration of RP30-R primer is 8pmol, the concentration...
Embodiment 3
[0112] Embodiment 3 Sensitivity detection
[0113] Connect the target fragment into the constructed pET28a-MS2 vector and transform the expression host strain BL21 competent cells, pick a single clone and sequence it for verification, then induce expression, and digest the completely fragmented expression product with RNaseA and DNaseⅠ. Virus-like particles containing fragments of interest.
[0114] Dilute the pseudovirus after the concentration determination to an appropriate concentration and then perform 10-fold dilution, the concentrations are 1.00E+06, 1.00E+05, 1.00E+04, 1.00E+03, 1.00E+02copies / ml.
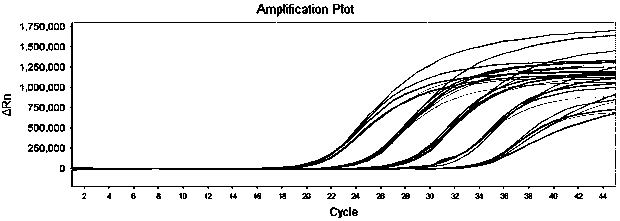
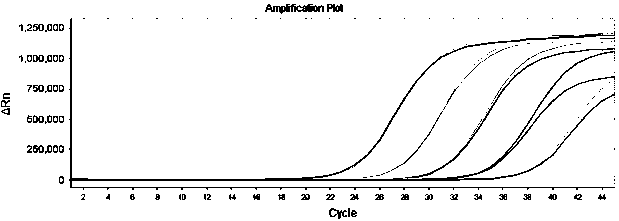
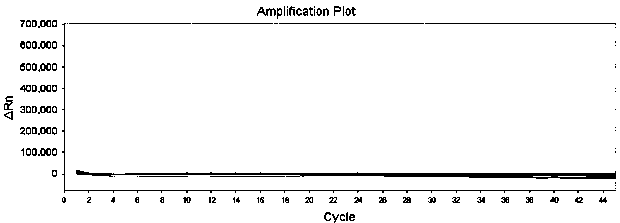
[0115] The above-mentioned pseudovirus is detected with the above-mentioned determined detection system and cycle parameters ( figure 2 ). The results show that this detection method has high sensitivity, the sensitivity is 1.00E+02 copies / ml.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com