Protein lysine malonylation site prediction method based on deep learning
A technology of lysine malonylation site and malonylation site, which is applied in the field of biological information, can solve the problems of reducing prediction performance, influence of prediction results, etc., and achieves the effect of improving evaluation index and promoting application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
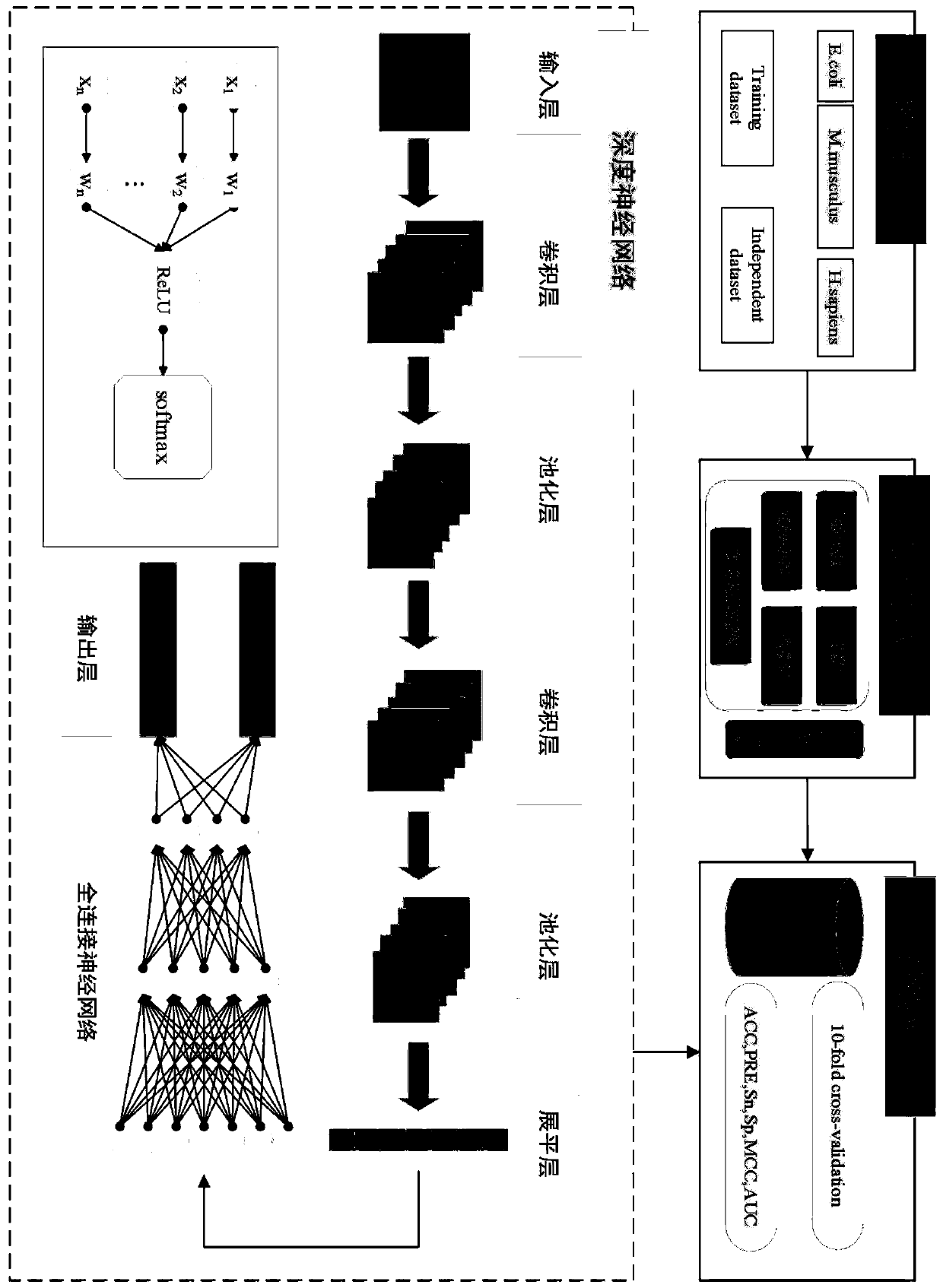
[0053] A method for predicting protein lysine malonylation sites based on deep learning, such as figure 1 shown, including the following steps:
[0054] 1) Data collection: Collect experimentally validated lysine malonylation site data from protein databases and related literature.
[0055] The experimentally verified lysine malonylation data set used in the present invention mainly comes from papers (Zhang YJ, XieRP, Wang JW, et al. Computational analysis and prediction of lysine malonylationsites by exploiting informative features in an integrative machine-learningframework. BriefBioinform 2018:1-15), this dataset includes 1746 Kmal sites from 595 E. coli proteins, 3435 Kmal sites from 1174 proteins from M. musculus and 4579 Kmal sites from 1660 proteins in H. sapiens.
[0056] After random selection, the final training set E.coli contains 1453 positive samples and 1453 negative samples, M.musculus contains 2606 positive samples and 2606 negative samples, and H.sapiens con...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com