Organochlorine pesticide ddt nucleic acid aptamer and its application
A nucleic acid aptamer and organochlorine technology, applied in the biological field, can solve problems such as complex processing, expensive instruments, and inability to analyze and detect a large number of samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1D
[0034] Example 1 Screening of DDT nucleic acid aptamers
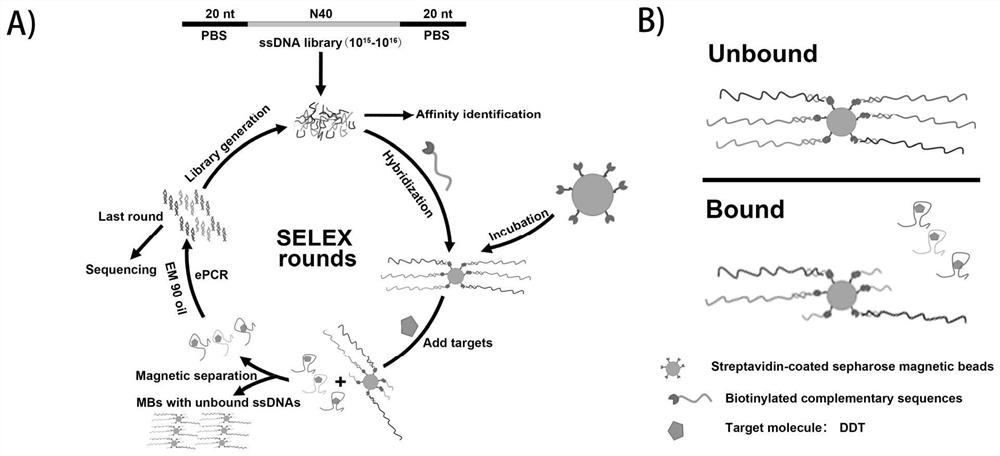
[0035] like figure 1 As shown, the present embodiment adopts SELEX technology, by fixing the library, and using the method of target elution to carry out nucleic acid aptamer screening, as follows:
[0036] 1. Immobilization of nucleic acid library and matrix (SA agarose magnetic beads)
[0037] The ssDNA nucleic acid aptamer library synthesized by Sanko is used, and the library information is 5'-TCCAGCACTCCACGCATAAC(SEQ ID NO.18)(Nn)GTTATGCGTGCGACGGTGAA(SEQ ID NO.19)-3', wherein n=20-60, preferably n=40; for the first round of screening, the 1OD library was dissolved in 260 μL Binding buffer (1 mM CaCl 2 , 2.5mM KCl, 1.5mM KH 2 PO 4 , 0.5mM MgCl 2 *6H 2 O, 137mM NaCl, 8mM Na 2 HPO 4 , pH 7.2), followed by annealing hybridization with 2-fold concentration of the upstream primer containing Biotin modification (5'-GTTATGCGTGGAGTGCTGGA-C6-Biotin-3', SEQ ID NO.20), the annealing procedure was 95°C for 10min; then...
Embodiment 2
[0052] Example 2 Cloning and sequencing of nucleic acid aptamers and prediction of secondary structure of candidate nucleic acid aptamers
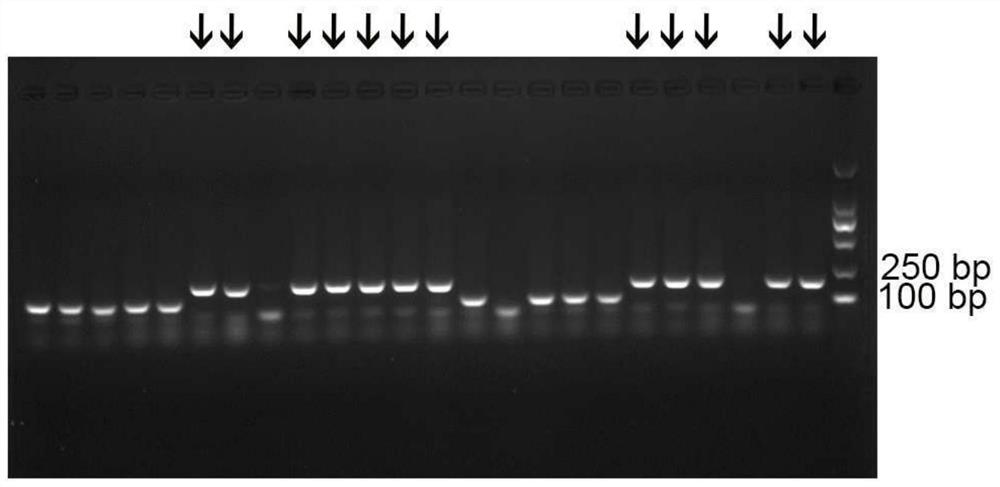
[0053] 1. Recovery of PCR products
[0054] Unmodified upstream and downstream primers were used to amplify the last round of screening in Example 1, followed by non-denaturing gel separation, cutting the gel to recover the target band, and using the ploy-gel gel recovery kit to recover the target product.
[0055] 2. Conversion of ligation and ligation products
[0056] 1. Add 0.5μL TAKARA pMDT-19T simple vector, 4.5μL nucleic acid aptamer PCR product and 5μL Solution I to a microcentrifuge tube;
[0057] 2. Reaction at 16°C for 3h;
[0058] 3. Add the full amount (10 μL) to 100 μL of DH5a competent cells, and place in ice for 30 min;
[0059] 4. After heat shock at 42°C for 90s, place on ice for 2min;
[0060] 5. Add 800 μL of pre-warmed LB medium at 37°C (without resistance), and incubate at 37°C with shaking at 220 RPM for 60 minut...
Embodiment 3
[0068] Example 3: Preliminary characterization of nucleic acid aptamers and determination of dissociation constant (Kd)
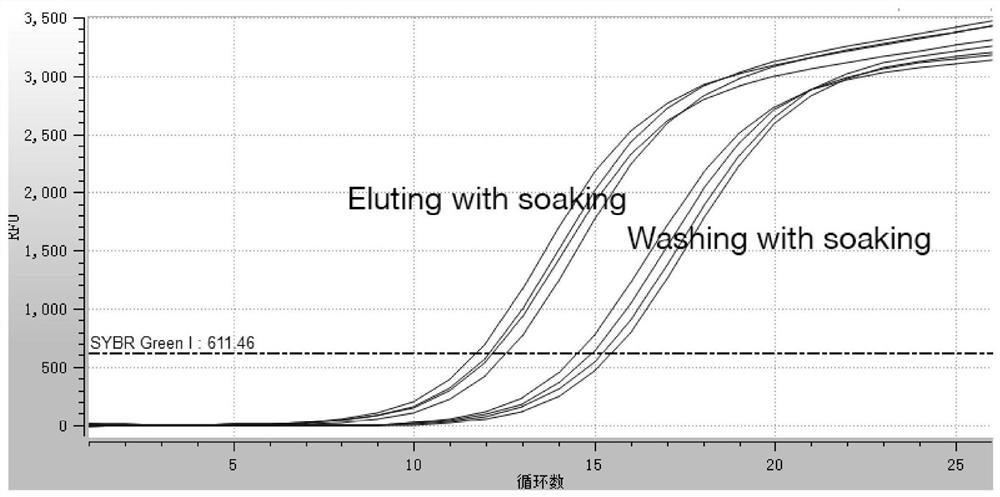
[0069] Using SYBR Green I (SGI) dye method, this method mainly uses nucleic acid aptamer to increase its secondary structure to a certain extent after binding to the target, thereby causing the continuous increase of SGI signal.
[0070] 1. Incubation of aptamer and target
[0071] The aptamers were incubated with the same concentration (200 nM) or different concentrations of the target (0-530 nM) for 45 min on a circular mixer.
[0072] 2. Incubation of aptamer-target complexes with SGI
[0073] The aptamer-target complex and SGI (final concentration of 1x) were incubated circumferentially, room temperature, for 2 h.
[0074] 3. Determination by fluorescence spectrophotometer
[0075] Fluorescence photometry was performed using a Hitachi F-7000 fluorescence spectrophotometer. Excitation wavelength EX 495; emission wavelength Em 505; slit excitation and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com