Molecular marker related with cashmere fineness trait and detection primer and application of molecular marker related with cashmere fineness trait
A technology for molecular labeling and detection of primers, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problem of undiscovered cashmere fineness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
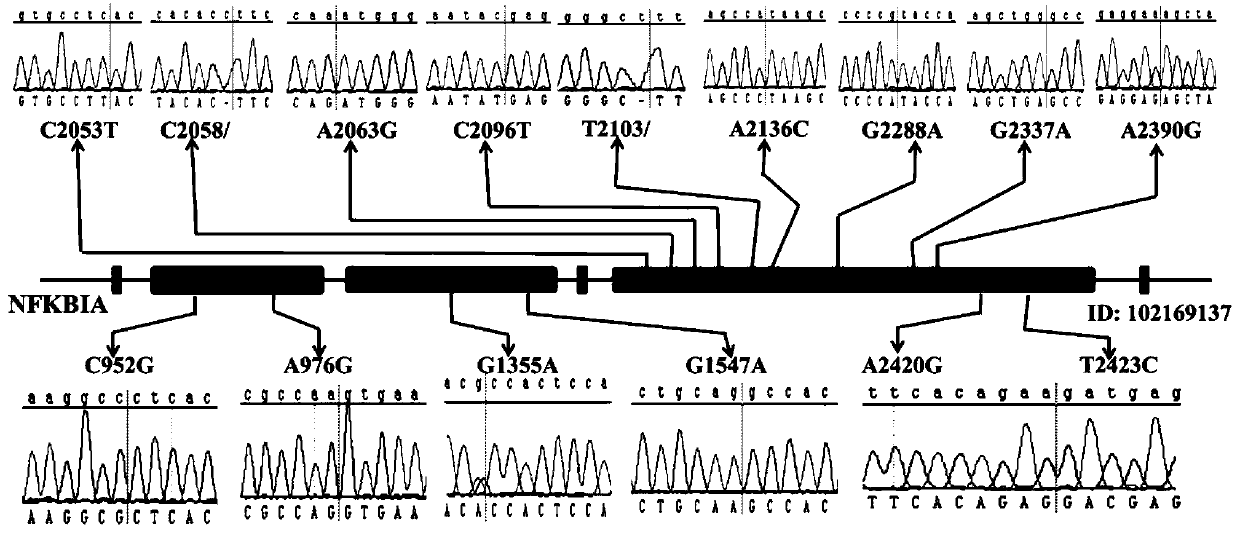
[0023] This embodiment provides a molecular marker related to cashmere fineness traits, which includes the G1355A site of the cashmere goat NFK BIA gene; the nucleotide sequence of the G1355A site is shown in the sequence table SEQ ID NO:1, Specifically: ACRCCACTCCACTTGGCTG; wherein, R in the nucleotide sequence of the G1355A site is A or G; the AG genotype at the G1355A site can be used for the breeding of fine cashmere goats.
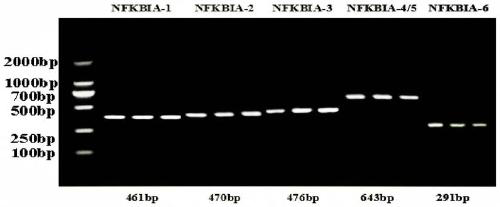
[0024] In addition, this embodiment also provides a detection primer for detecting the above-mentioned molecular markers, specifically, the detection primers include five sets of primer pairs, specifically, the detection primers are used to detect the Molecular marker; the detection primers include at least one of NFKBIA-1 primer pair, NFKBIA-2 primer pair, NFKBIA-3 primer pair, NFKBIA-4 / 5 primer pair and NFKBIA-6 primer pair; the NFKBIA- 1 The nucleotide sequence of the primer pair is shown in the sequence table SEQ ID NO: 2~3; the nucleotide sequenc...
Embodiment 2
[0029] This embodiment provides a method for detecting the mutation site in the cashmere goat NFKBIA gene using the above-mentioned detection primers, comprising the following steps:
[0030] S1. Obtain the DNA of the cashmere goat to be detected to obtain a DNA template;
[0031] S2. Perform PCR amplification on the detection primer and the DNA template to obtain a PCR amplification product;
[0032] S3. Sequencing the PCR amplification product to obtain a sequenced gene sequence;
[0033] S4. Comparing the sequenced gene sequence with the cashmere goat NFKBIA standard gene sequence to obtain the mutation site in the cashmere goat NFKBIA gene to be detected.
[0034] Concretely, in actual application, the above-mentioned method can be used in the experiment of finding the marker genotype related to the fineness economic traits of Liaoning cashmere goat cashmere, and its experimental method can include the following steps:
[0035](1) At the cashmere goat breeding farm in Li...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com