A molecular marker method and application of peanut flowering habit-related gene loci
A technology of flowering habit and gene locus, which is applied in the field of molecular genetics and molecular breeding of crops to save costs, improve the efficiency of breeding, and speed up the breeding speed.
Active Publication Date: 2022-05-31
QINGDAO AGRI UNIV
View PDF3 Cites 0 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0005] Existing studies have not found a unified understanding, and there is no more accurate positioning of the genetic loci related to peanut flowering habit and the development of molecular marker-assisted selection markers
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment 1
[0049] (2) using the improved CTAB method to extract the population genomic DNA (specifically refer to "Molecular Cloning Experiment Guide").
[0052]
Embodiment 2
[0071] The PCR reaction system is 10 μl, of which 12.5 μl of 2×Taq Master Mix for PAGE, 1 μl of template, 10 μM
Embodiment 3
[0079] (2) Extract the population genomic DNA with the improved CTAB method (specifically refer to the "Molecular Cloning Experiment Guide").
[0084]
[0085]
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
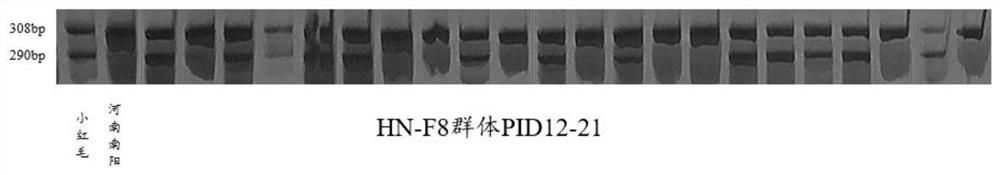
The invention belongs to the technical field of crop molecular genetics and molecular breeding, and provides a molecular marker method and application of peanut flowering habit-related sites. The molecular markers include InDel markers PID12‑21 and PID12‑29 located on the 12th The InDel marker PID2‑9 of chromosome 02; the flowering habit of the single-gene inheritance hybrid population only needs to consider the genotype of the locus related to the flowering habit of chromosome 12; The genotype of the locus related to the flowering habit of the chromosome; whether it is a hybrid population with a single or double gene inheritance model, only the genotype of the amplified target band can be identified to distinguish the continuous flowering type and the alternate flowering type of peanuts, Quickly screen the types of flowering habits of peanut hybrid offspring, and assist in plant type breeding and yield breeding related to flowering habits.
Description
A molecular marker method for peanut flowering habit related gene loci and its application Technical field: The invention belongs to crop molecular genetics and molecular breeding technical field, relate to peanut flowering habit related gene locus Point-based molecular marker-assisted selection method for molecular marker-assisted improvement of peanut flowering habit-related traits, further To achieve molecular marker-assisted improvement of peanut branch number, plant type and yield. Background technique: Peanut (Arachis hypogaea L.) is one of the important oil crops in my country, and it is rich in many Nutrients such as unsaturated fatty acids, protein and trace elements. Cultivated peanut (allotetraploid AABB, 2n=4x= 40) was domesticated by natural outcross doubling of two diploid wild species with different genome types, and their genome structures are similar. For complex, genetic studies were carried out relatively late. With the current whole genome seq...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Patents(China)
IPC IPC(8): C12Q1/6895C12N15/11
CPCC12Q1/6895C12Q2600/13C12Q2600/156Y02A40/146
Inventor 张晓军李季华郭蕊于晓娜司彤邹晓霞王月福王铭伦
Owner QINGDAO AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com