SNP locus, primer group and method for identifying cauliflower variety Zhejiang Nongsong 80-day
A cauliflower and primer set technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve problems such as long investigation cycle, achieve high practical value, accurate identification, and shorten the identification cycle effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
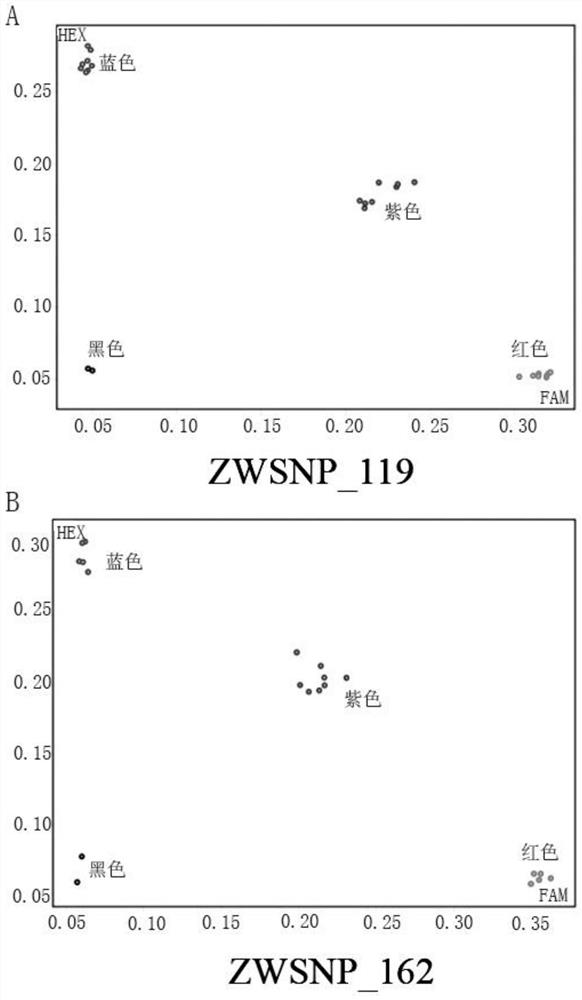
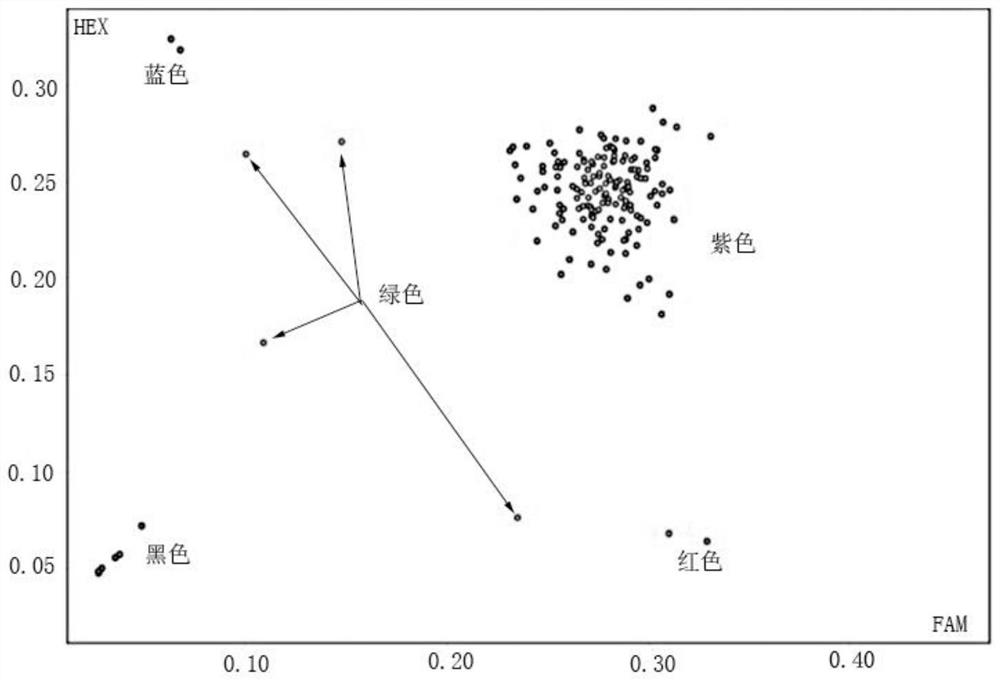
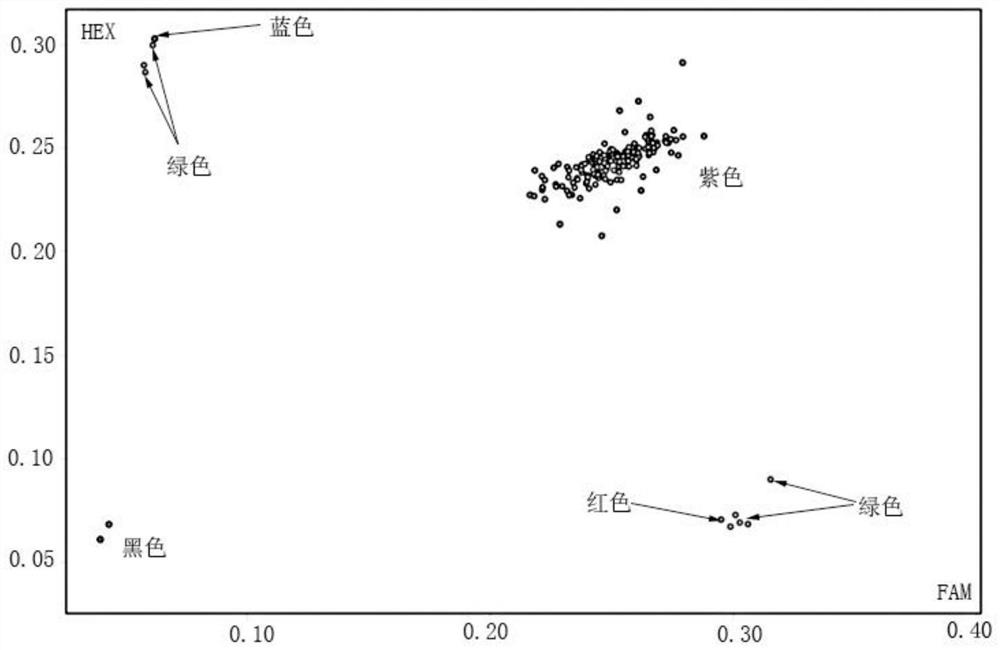
[0045] A method for identifying the purity of cauliflower "Zhenong Songhua 80 days" by KASP labeling, using the above-mentioned KASP labeling primer set, adding a specific sequence that can bind to FAM fluorescence at the 5' end of SNP-specific primers X and X' (SEQ ID. NO 7: GAAGGTGACCAAGTTCATGCT), the 5' end of the SNP-specific primer Y and Y' added a specific sequence that can be combined with HEX fluorescence (SEQ ID. NO 8: GAAGGTCGGAGTCAACGGATT), the identified materials are: "Zhenong Songhua 80 days" 2 male parents, 2 female parents and samples. The samples are 98 true hybrids of "Zhenong Songhua 80 days" and 4 artificial hybrid materials. The method is as follows:
[0046] (1) Extraction of genomic DNA
[0047] Genomic DNA was extracted from the “Zhenong Songhua 80 Days” hybrid and its parents using the traditional CTAB method, as follows:
[0048] Take 0.1 g of the young leaves of the plants to be selected, put them into a 2.0 mL flat-top centrifuge tube, add 600 μL o...
Embodiment 2
[0057] Authenticity identification of hybrids harvested in the field of embodiment 2
[0058] "Zhe Nong Songhua 80 days" has 1 male parent and 1 female parent, and 208 hybrids were harvested that year. These materials were raised in plug trays on July 15, 2019, and transplanted into the field on August 15, 2019. From October 15 to November 15, 2019, the agronomic traits such as leaf shape, plant shape and ball shape of 208 materials were tested. Observe, and compare with the parent material of father and mother, determine the number of hybrids and the purity of the sample to be tested, and identify by the method of Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com