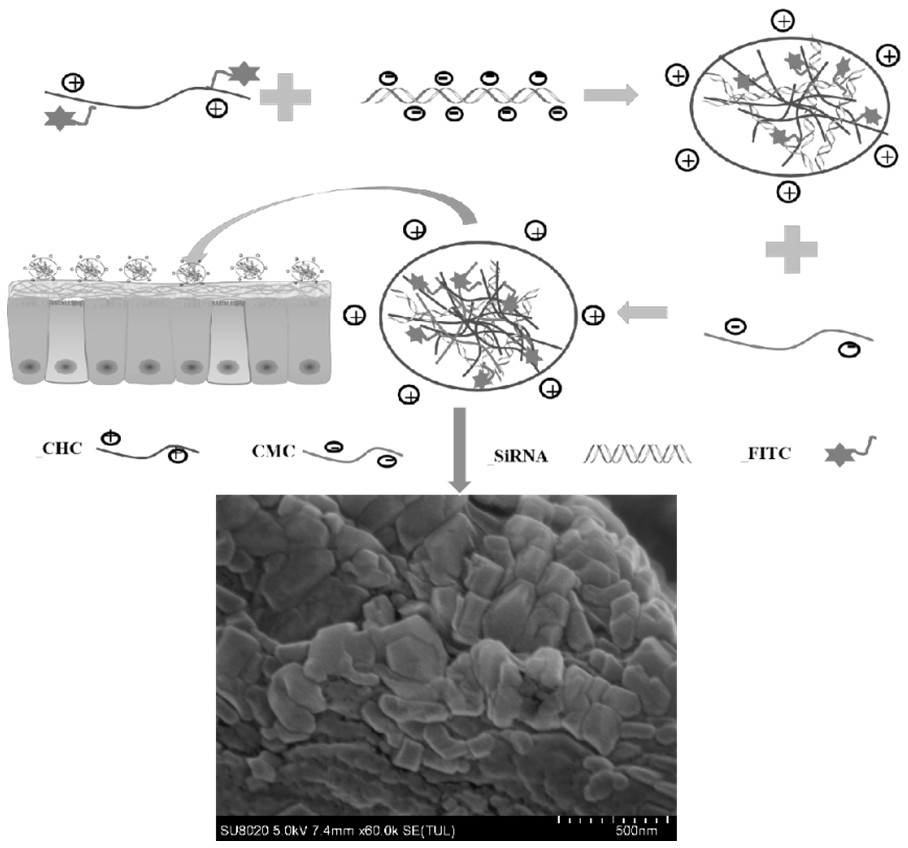
A preparation method of chitosan derivative nanoparticles delivering siRNA
A chitosan derivative and nanoparticle technology, applied in the field of gene biology, to achieve the effect of promoting the release of siRNA, inhibiting proliferation and migration, and the method is simple
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] The concrete operating steps of the chitosan hydrochloride powder of preparation FITC mark are as follows:
[0026] 10 mg chitosan hydrochloride was completely dissolved in 10 mg deionized water to obtain a chitosan hydrochloride solution with a concentration of 1 mg / mL. Add 10 mL of fluorescein isothiocyanate (FITC) solution (5.8 mg / L, anhydrous methanol) with a concentration of 5.8 mg / L to the chitosan hydrochloride solution with a concentration of 1 mg / mL, and stir under magnetic stirring (500 rpm ) for 4 h. Then add 1 M concentration of hydroxide (NaOH) to adjust the pH value to 10, and precipitate FITC-labeled chitosan hydrochloride. The free FITC was washed with ultrapure water and centrifuged until no fluorescence was detected in the supernatant. Finally, FITC-labeled chitosan hydrochloride powder was obtained by freeze-drying.
[0027] Prepare siRNA-CDNPs according to siRNA in Table 1, different volumes of CHC and different volumes of CMC (600 μL, 500 μL, 400...
Embodiment 2
[0033] Application of siRNA-CDNPs in the controlled release of siRNA in a simulated environment of colon cancer cells:
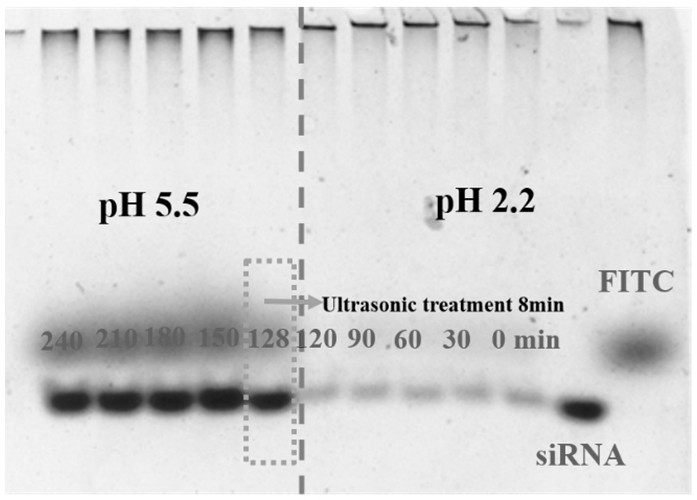
[0034]The siRNA-CDNPs prepared in Example 1 were incubated in a gastric environment (pH value = 2.2, 120 rpm) at 37°C for different times, up to 120 min. The above acidic solution was then mixed with NaOH (1 M) to adjust the pH to a weak acid (pH = 5.5). Then sonicate for 8 min, incubate at 37°C, and shake well (120 rpm) for 112 min. Samples were taken at predetermined times (0, 30, 60, 90, 120, 128, 150, 180, 210 and 240 min). Samples were analyzed on 5% (w / v) polyacrylamide gel, electrophoresis was performed at 120 mV in 1× TBE buffer, the gel was stained with ethidium bromide, and observed with a gel imager FLA 7000, the results were as follows figure 2 shown.
[0035] The release of siRNA from siRNA-CDNPs was detected by polyacrylamide gel electrophoresis by analyzing the siRNA content at pH 2.2 and pH 5.5. figure 2 The red and white arrows in ind...
Embodiment 3
[0037] Application of siRNA in siRNA-CDNPs efficiently internalized by HT-29 cells:
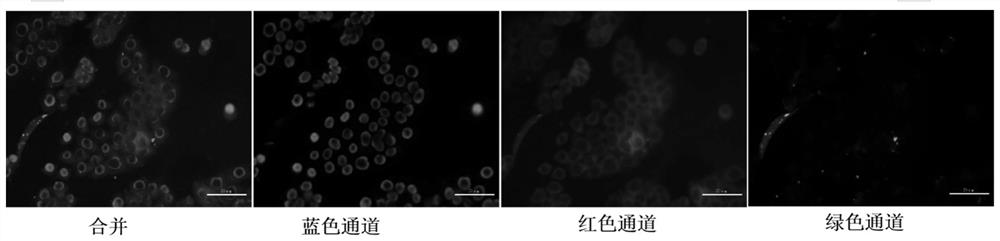
[0038] Incubate colon cancer cells (HT-29) with siRNA-CDNPs prepared in Example 1, and analyze the transfection effect of siRNA. The specific steps are as follows: Add 100 μL siRNA-CDNPs (containing 0.55 nmol siRNA) to HT-29 cells (1×10 5 / well) for 16 h. The added siRNA-CDNPs were then gently absorbed, and the cells were washed three times with PBS. Cells were fixed with 4% paraformaldehyde in PBS for 15 min, washed twice with PBS, and nuclear stained with DAPI (4′,6-diamino-2-phenylindole) for 10 min. Cells were washed again with PBS, rinsed with ultrapure water, and placed on microscope slides. Fluorescence images were taken with a fluorescence microscope, and the results were as follows image 3 shown.
[0039] The siRNA-CDNPs prepared in Example 1 were added to colon cancer cells (HT-29), and the transfection effect of siRNA was analyzed. The results were as follows: image 3 shown...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com