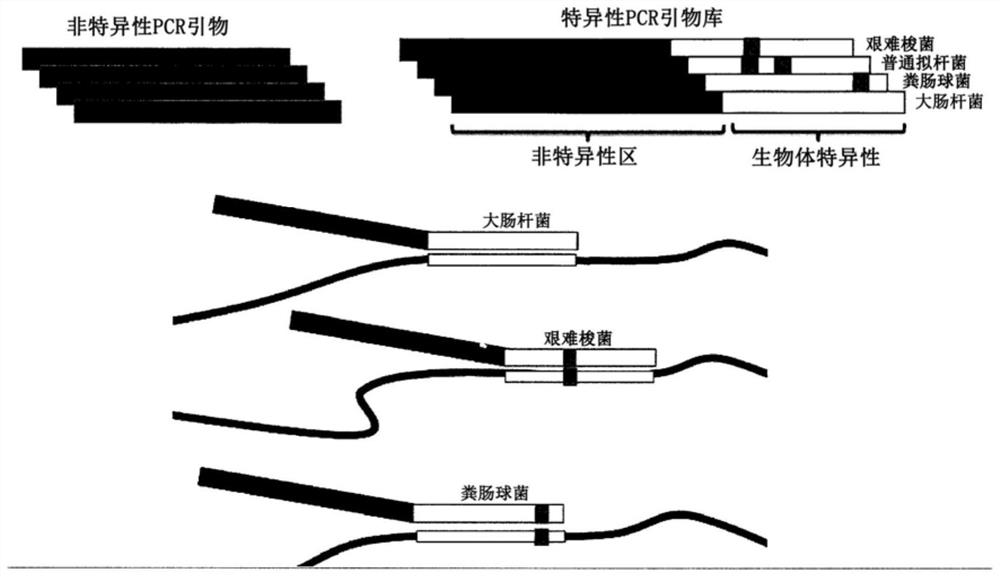
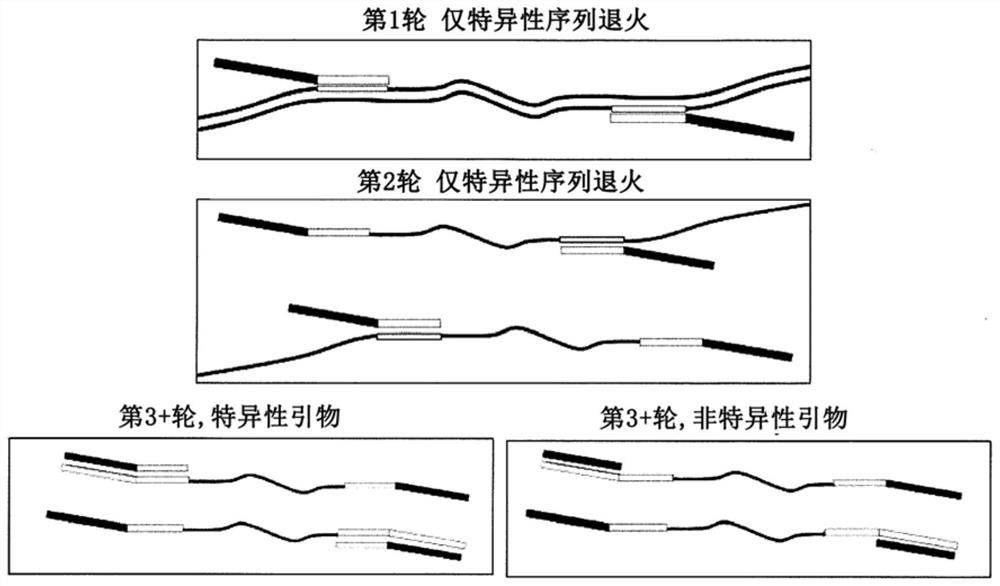
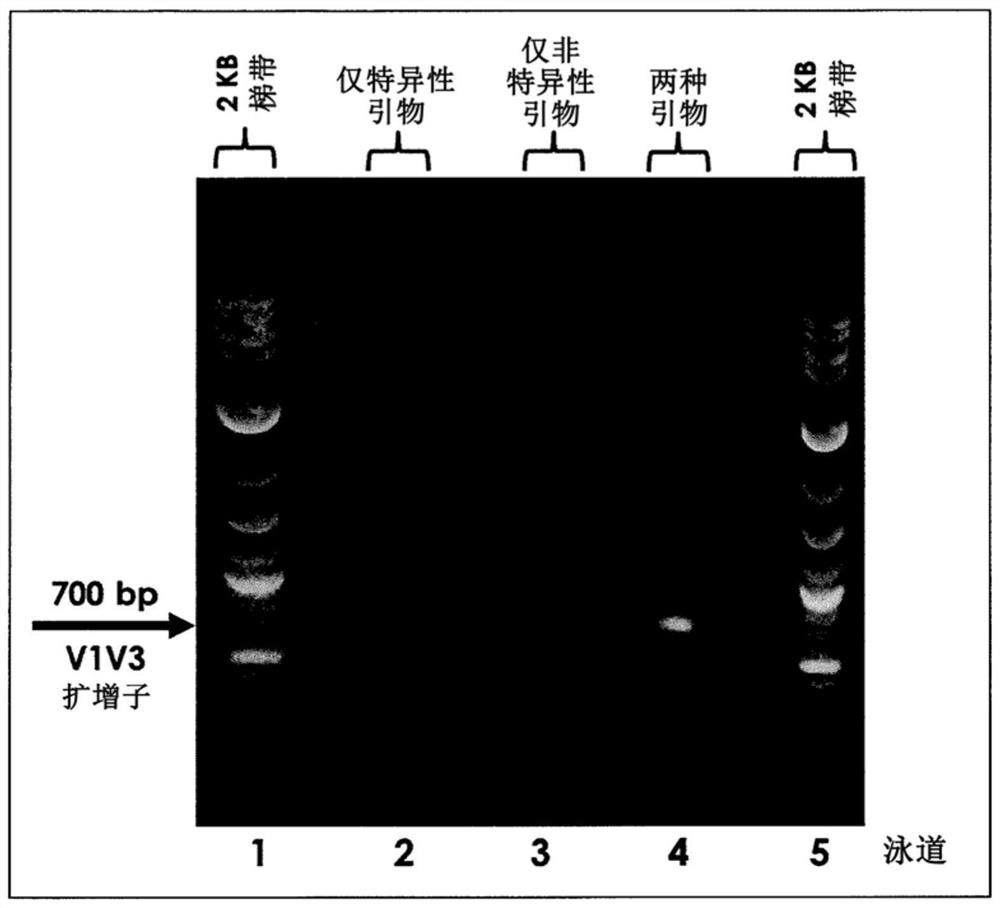
Multiple specific/nonspecific primers for PCR of a complex gene pool
A non-specific and specific technology, applied in the field of multiple specific/non-specific primers for PCR of complex gene pools, which can solve the complex interpretation of microbiome data, distortion of the original population ratio of target genes, increase in low abundance high abundance, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0026] The examples described herein apply the compositions and methods of the invention to the amplification of DNA sequences from microbiome samples. DNA was generated by lysing cells in the target microbiome, and the resulting DNA was used as template in PCR amplification targeting the 16S rRNA gene present in all bacteria and archaea. Microorganisms can be identified using their 16S rRNA gene sequence, which is slightly different in most, if not all, bacteria and archaea. Variation in the 16S gene sequence means that individual species of bacteria and archaea have characteristic DNA variations in the 16S rRNA gene that serve as an identifier or fingerprint for that species. Kits, protocols, and software available from Shoreline Biome (Farmington, CT) enable comprehensive fingerprinting of microorganisms in a sample using amplicons engineered in both the 16S rRNA and 23S rRNA genes, and simultaneously, at high-resolution Perform 16S rRNA analysis on many samples at once. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com