Quantitative 16S metagenome sequencing method
A metagenomic, 16SV1-V3 technology, applied in the field of quantitative 16S metagenomic sequencing, can solve problems such as poor comparability, data cannot be directly applied to research topics and testing projects, and cannot be compared, etc., to reduce impact, have horizontal comparability, The effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
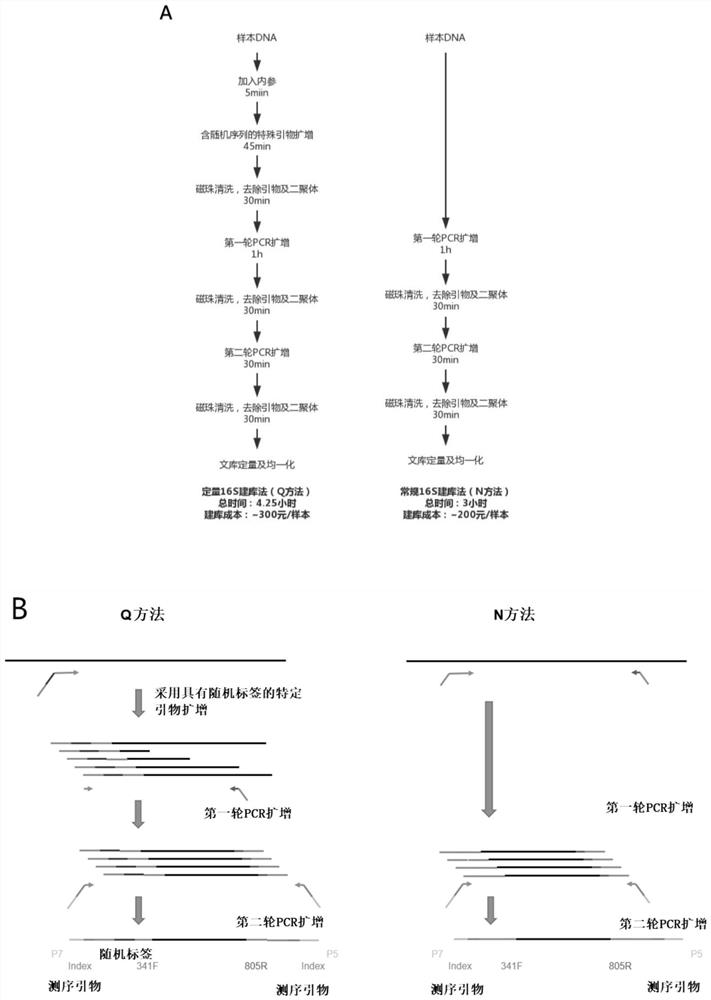
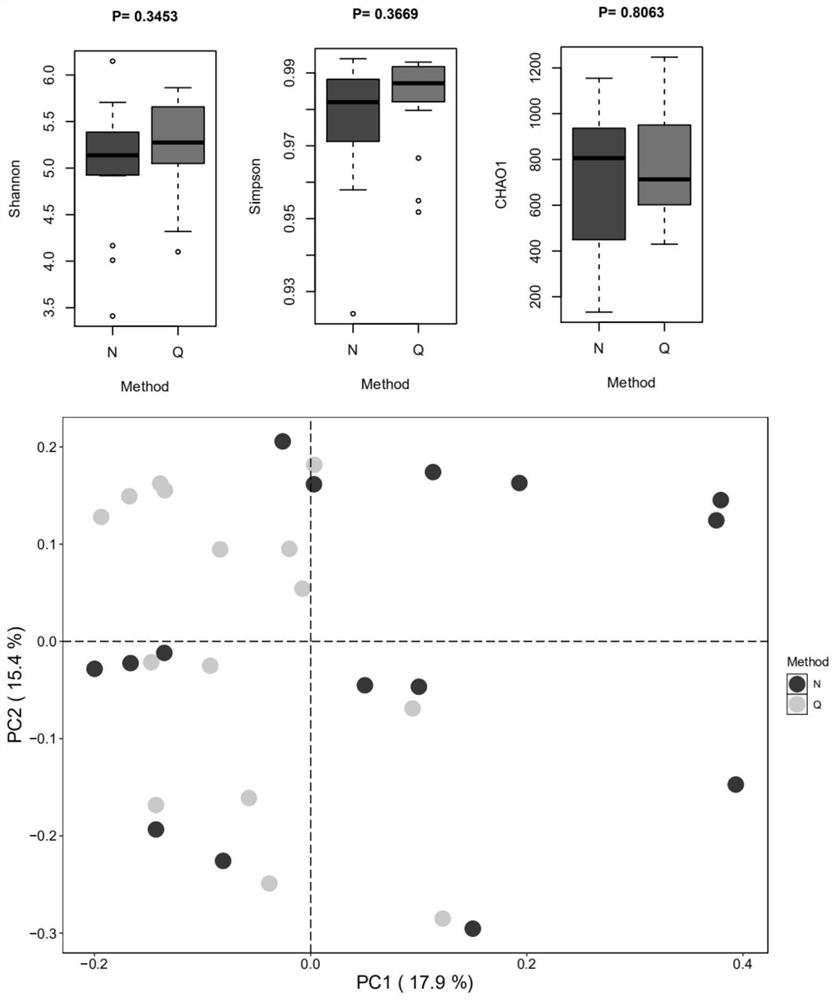
[0061] Example 1 Comparison of Quantitative 16S Metagenome Sequencing Method (Q Method) and Conventional 16S Sequencing Method (N Method)
[0062] 1. Internal reference design
[0063] Two plasmids containing inserted sequences were artificially designed as internal references for sequencing (internal references). The internal reference sequence simulates the 16S V3 / V4 region, and its structure is 341F amplification primer+random sequence (300bp)+805R primer. The inserted random sequence has been verified by the Blast method, and there is no match with known natural species sequences. According to the synthesized plasmid concentration and molecular weight, the number of plasmid copies can be converted. Add 50,000 copies of internal reference 1 and 10,000 copies of internal reference 2 as internal references when building a library for each sample.
[0064] 2. Sampling and sequencing experiments
[0065] Questionnaires were used to investigate the volunteers' personal condi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com