Marker, detection method and detection system for homologous recombination deletion
A homologous recombination and marker technology, applied in the field of markers lacking homologous recombination, can solve the problems of low sequencing depth, low number of heterozygous SNPs, and inability to detect low-abundance somatic mutations, and achieve consistency Good results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
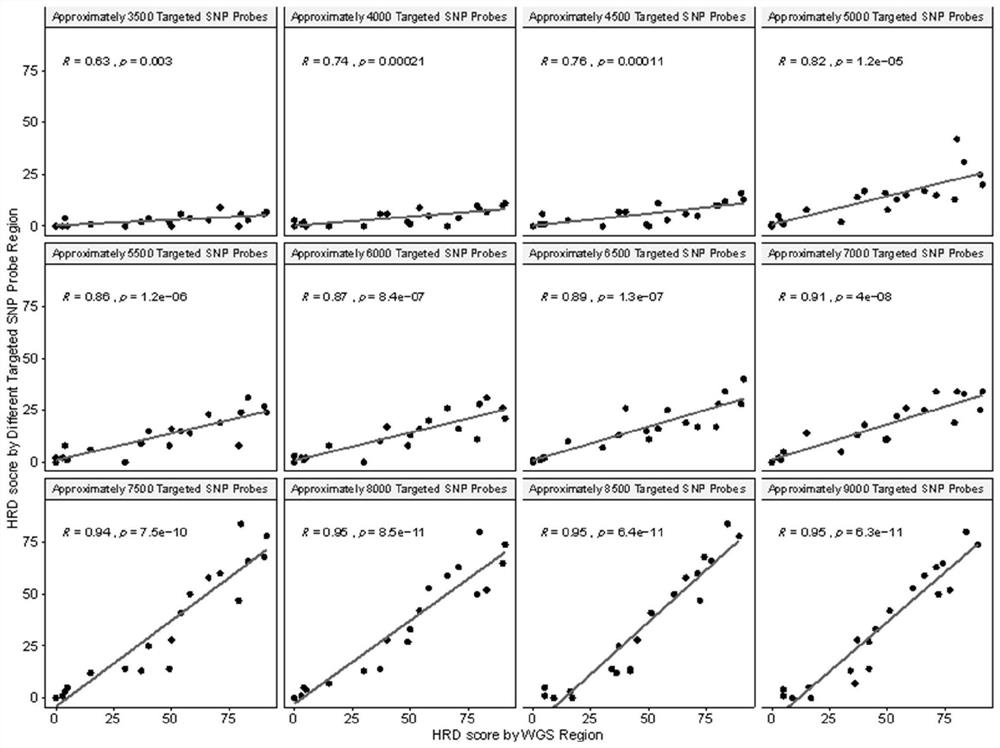
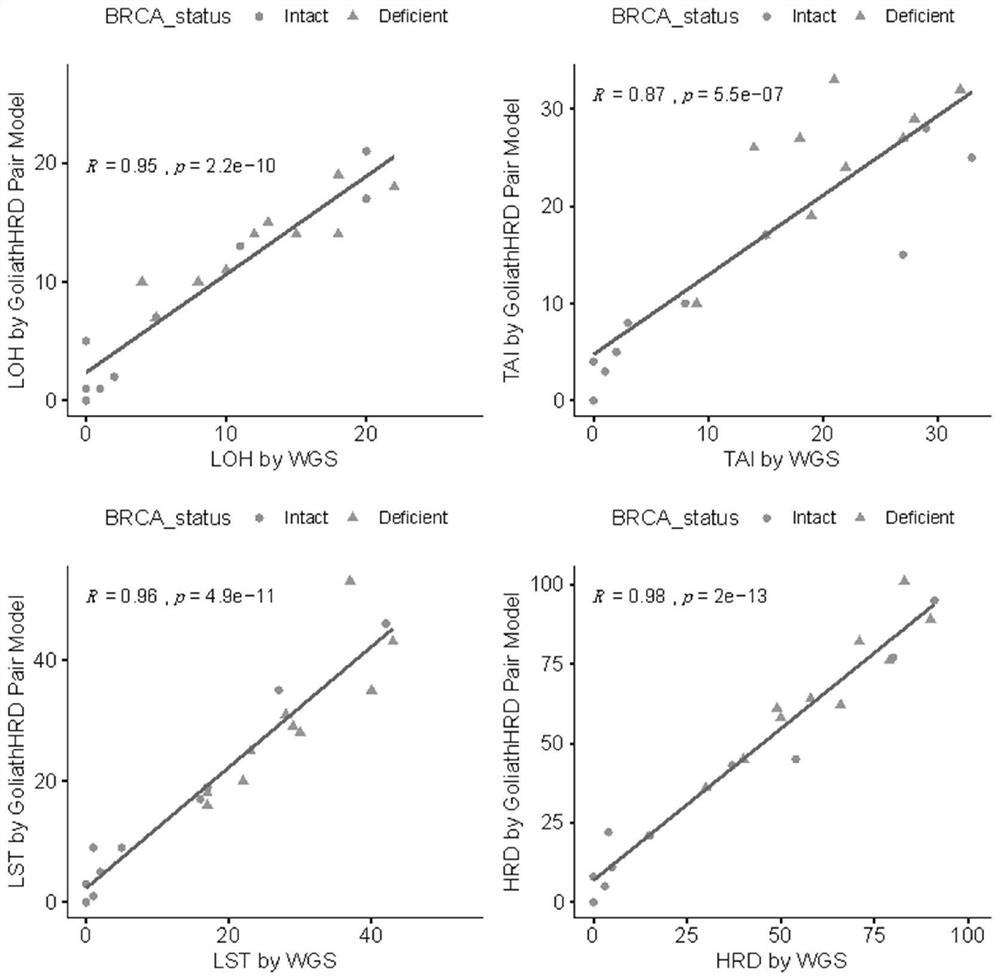
[0046] Loss of heterozygosity is often found in cancer, and the B allele mutation frequency (BAF) of the chromosomal region in the region of the loss of heterozygosity changes, as shown in the figure, in this region BAF = 50% in healthy samples, while Due to the occurrence of heterozygous deletion in tumor tissue, BAF=100%, so in order to better detect homologous recombination deletion, a large number of heterozygous SNP sites were designed in the panel.
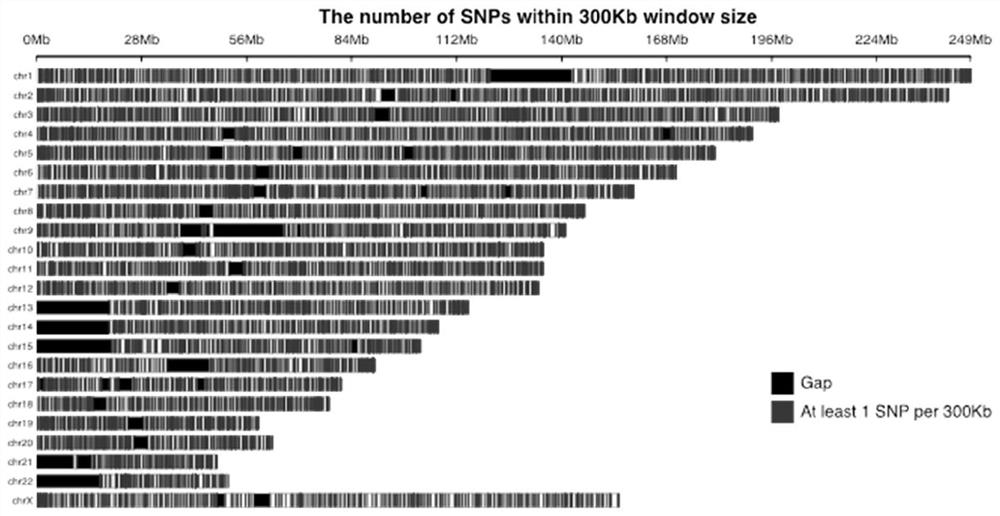
[0047] In the present invention, the most common polymorphic sites based on Asians are screened, and NGS sequencing is performed through the above-mentioned panel composed of multiple SNP site regions spanning the entire genome. combined SNP loci. gnomAD v2.1 contains SNV and InDel results for 15,708 WGS samples, including 780 East Asian samples. Calculate the gnomAD site heterozygosity rate (gnomAD Het rate=2*EAS_AF*(1-EAS_AF)) according to the gnomAD East Asian population frequency (EAS_AF), and select the site with the h...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com