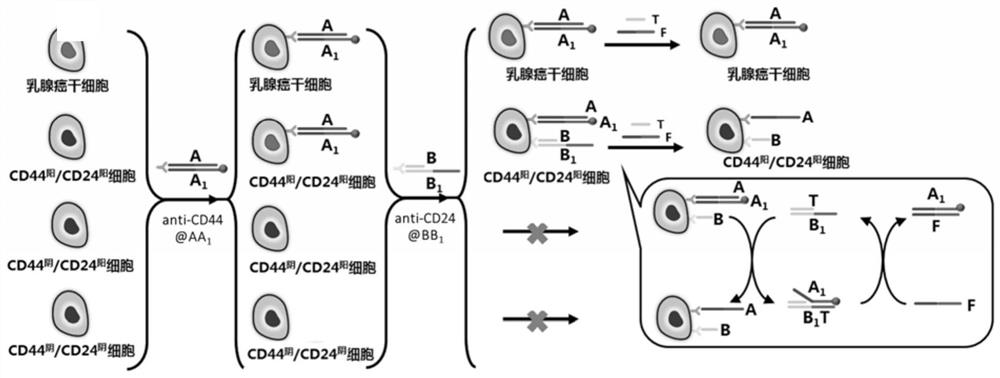
Method for identifying CD44 and CD24 molecular phenotypes
A CD24, CD44 technology, applied in the direction of analysis materials, measuring devices, instruments, etc., can solve the problems of large amount of data in double-color flow cytometry analysis, increase the workload and difficulty of data analysis, and dope with wrong information, etc., and meet the technical level requirements Low cost, easy operation, and small amount of data analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1 Electrochemical signal labeling of breast cancer stem cells based on CD44 antibody-nucleic acid conjugates
[0058] (a) Take 20 μL of DNA probes A and A at a concentration of 50 μM 1 Put it in a microtube, mix it evenly and put it under the condition of 25 ℃ for 2 hours to get AA 1 Double strand solution.
[0059] (b) Take 10 μL of the AA prepared in step (a) 1 The double-strand solution was placed in a microtube, and then 10 μL of 30 mM ascorbic acid solution, 10 μL of 2 mM copper sulfate solution, and 70 μL of 3-(N-morpholino)propanesulfonic acid (MOPS) buffer ( Containing 20mM MOPS, 300mM NaCl, pH 7.5), reacted at 25°C for 15 minutes, so that A 1 The polycytosine sequence at the 5' end of the chain (T 30 ) as a template to synthesize copper nanoparticles that can be used as electrochemical signal probes.
[0060] (c) Dissolve 5 μL of CD44 antibody with a concentration of 1 μM and 5 μL of Tetrazine-PEG5-NHS with a concentration of 5 μM in 100 μL of phos...
Embodiment 2
[0067] Electrochemical analysis of embodiment 2 breast cancer stem cells
[0068] (a) Take 20 μL of DNA probes A and A at a concentration of 50 μM 1 Put it in a microtube, mix it evenly and put it under the condition of 25 ℃ for 2 hours to get AA 1 Double-stranded solution; take 20 μL of DNA probes B and B at a concentration of 50 μM 1 Put it in a microtube, mix it evenly and put it at 25°C for 2 hours to get BB 1 Double-strand solution: Take 20 μL of DNA probes T and F with a concentration of 100 μM and place them in microtubes respectively to obtain T-strand and F-strand solutions.
[0069] (b) Take 10 μL of the AA prepared in step (a) 1 The double-strand solution was placed in a microtube, and then 10 μL of 30 mM ascorbic acid solution, 10 μL of 2 mM copper sulfate solution, and 70 μL of 3-(N-morpholino)propanesulfonic acid (MOPS) buffer ( Containing 20mM MOPS, 300mM NaCl, pH 7.5), reacted at 25°C for 15 minutes, so that A 1 The polycytosine sequence at the 5' end of t...
Embodiment 3
[0083] Electrochemical analysis of breast cancer stem cells under the coexistence condition of interference cells in embodiment 3
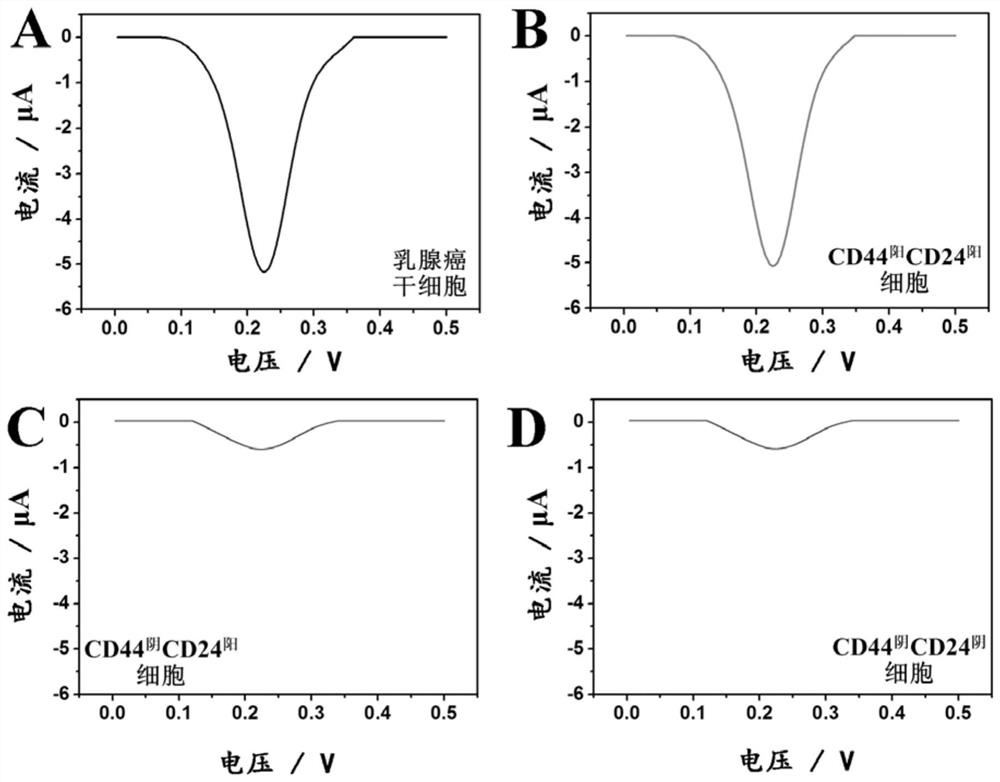
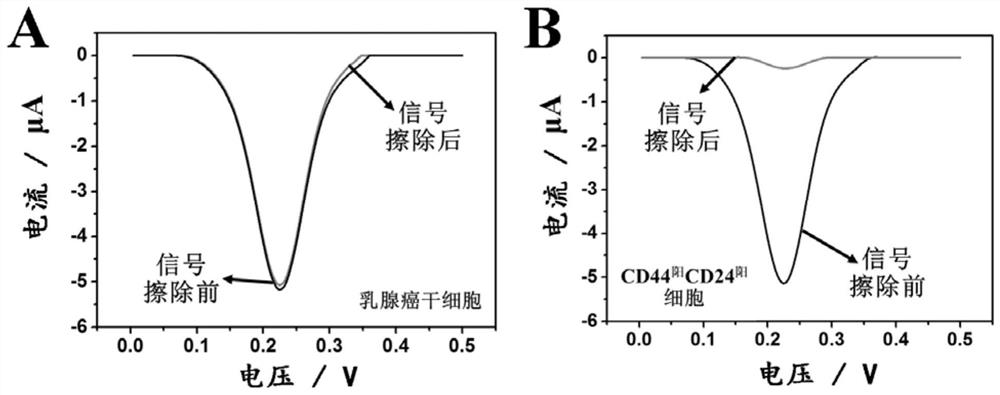
[0084] In order to examine the specificity and anti-interference ability of this technology for the analysis of breast cancer stem cells, we divided 5×10 5 pcs, 5×10 4 and 5×10 3 breast cancer stem cells with 5×10 5 Three different molecular phenotypes (CD44 阳 CD24 阳 , CD44 阴 CD24 阳 , CD44 阴 CD24 阴 ) of interfering cells were mixed, and electrochemical analysis was carried out according to the steps in Example 2. The result is as Figure 5 As shown, the presence of three different molecular phenotypes of interfering cells will not cause changes in the peak current value of the electrochemical response of breast cancer stem cells, indicating that this technology uses antibody-nucleic acid conjugate-based selective electrochemical signal labeling and The background signal erasing strategy based on programmable DNA circuit can effectively e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com