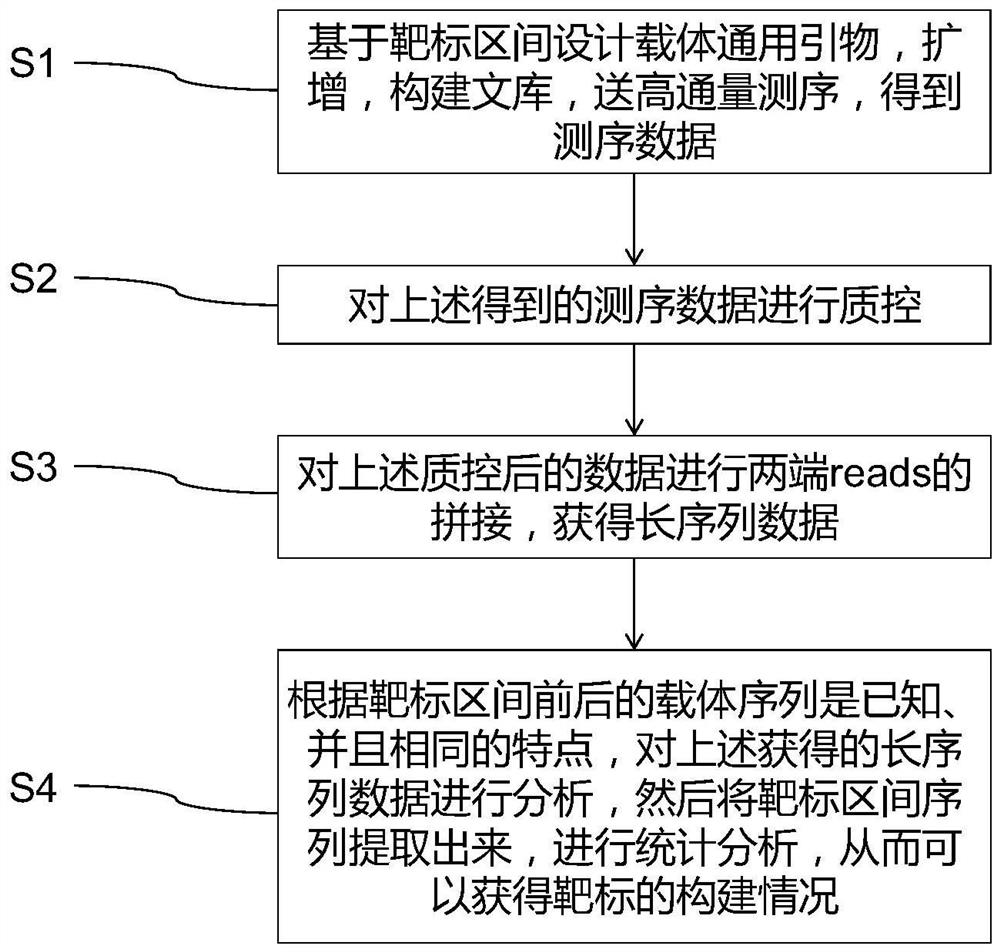
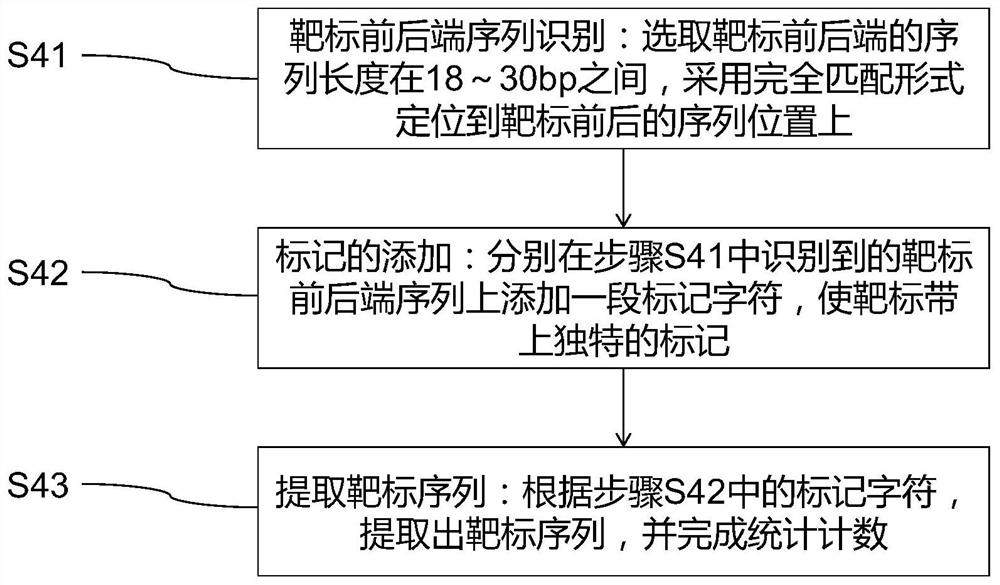
Method for rapidly analyzing construction condition of CRISPR/Cas9 gene editing vector and application
A technology of gene editing and rapid analysis, applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of high price and difficulty in large-scale use, and achieve the effect of low cost and good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] In the following example, 100 constructed CRISPR / Cas9 gene editing vectors were selected and mixed in equal proportions to form a gene editing vector library. The specific operation process is as follows:
[0036] 1.1 Primer design
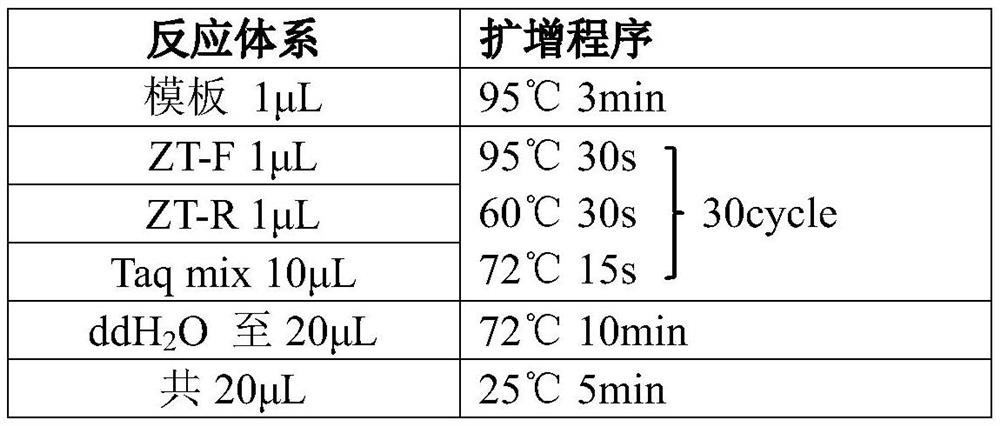
[0037] According to the vector sequence, design the universal primer sequence of the amplified vector as follows:
[0038] ZT-F: 5'-ggagtgagtaccgtgagcTGGACGACAACAAAGACTAG-3';
[0039] ZT-R: 5'-ggaatgcatgctgcatgcTGCCACTTTTTCAAGTTGAT-3';
[0040] The lowercase letters in the above universal primer sequences for vectors are bridge sequences (as shown in SEQ ID NO.1 and SEQ ID NO.2), and the uppercase letters are universal amplification primers for vectors (it should be noted that different gene editing vectors can be Design different vector universal primer sequences), this embodiment according to the sequencing method of PE150, the distance between the designed forward primer and reverse primer is 80bp from the target position.
[0041] 1.2 ...
Embodiment 2
[0060] Mix 100 successfully constructed CRISPR / Cas9 gene editing vector libraries into Agrobacterium, culture for 2 to 3 days, then scrape the Agrobacterium, resuspend all Agrobacterium, and infect the rice callus. After tissue culture, a large number of regenerated positive tissue culture seedlings were obtained, which were numbered according to the arrangement of the PCR 96-well plate: A1-H12.
[0061] 2.1 Primer design
[0062] According to the vector sequence, the primer design method in Example 1 was the same.
[0063] 2.2 Genome Extraction
[0064] Take the regenerated positive tissue-cultured seedlings, take leaves 1-2 cm in size, put them into a 96-deep well plate, add 50 μL of 0.2 M KOH solution and 1-2 grinding beads to each well, and centrifuge 2 mL of Transfer the tubes to the adapters of the fully automatic sample grinder and run for 30-60 seconds until the buffer turns green; then use a row gun to pipette 1 μL of the supernatant into a 96-well PCR plate as a te...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com