Polypeptide anticancer function identification method, system, medium and equipment
A recognition method and function technology, applied in the field of bioinformatics, can solve problems such as limiting prediction performance, reducing the recognition accuracy of polypeptide anti-cancer functions, affecting the robustness of prediction models, etc., and achieving the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
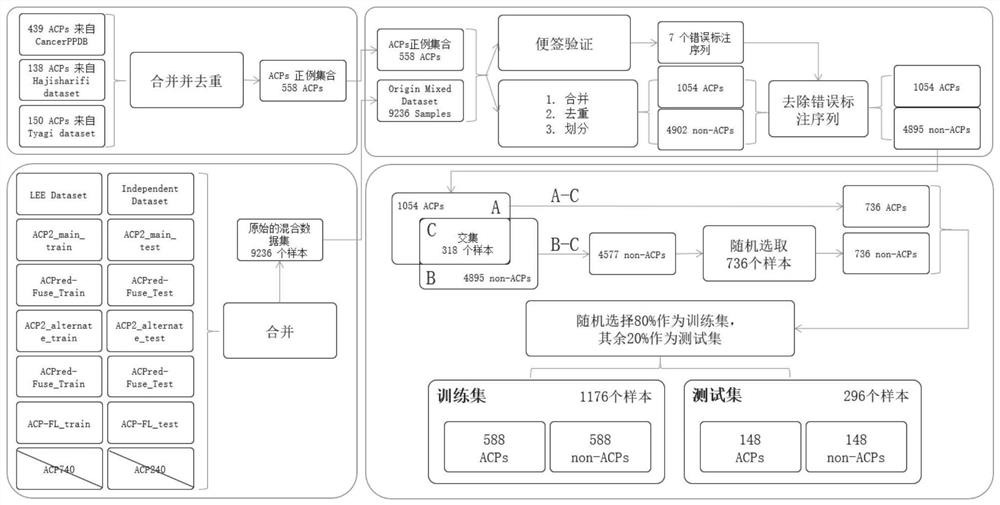
[0064] This embodiment provides a method for identifying the anti-cancer function of a polypeptide, which specifically includes the following steps:
[0065] Step S101: obtaining the polypeptide sequence;
[0066] Step S102: Input the polypeptide sequence into the trained polypeptide drug anti-cancer function predictor to obtain whether the polypeptide has anti-cancer function.
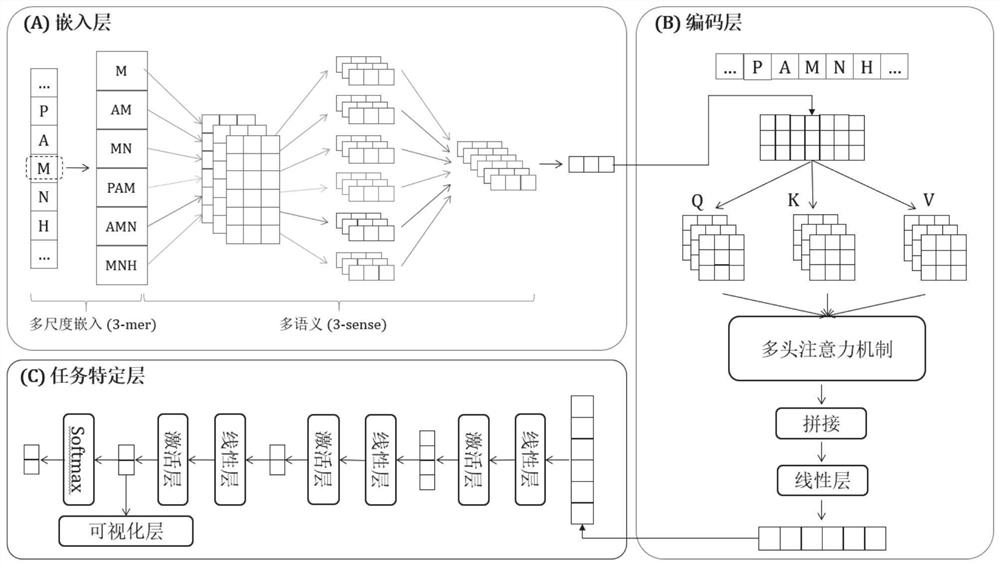
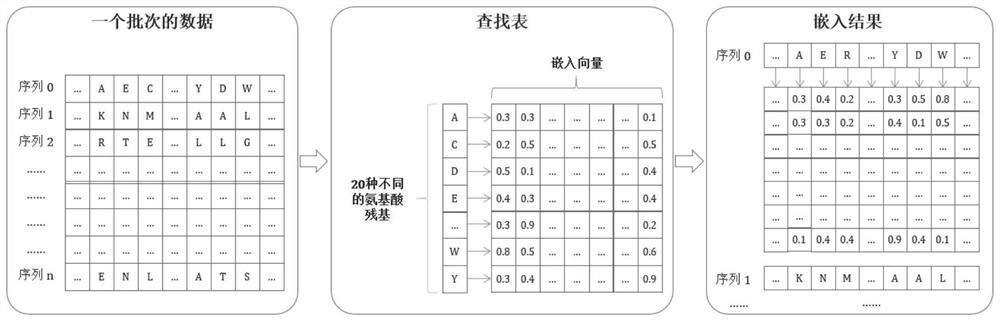
[0067] refer to figure 2 , the polypeptide drug anticancer function predictor of this embodiment includes an embedding layer, a coding layer and a task-specific layer, and the embedding layer is used to convert each residue of the received polypeptide sequence into a low-dimensional dense vector and in the form of a matrix Output; the encoding layer is used to capture the context of each residual embedding vector in different positions from the matrix output by the embedding layer, and learn the discriminative features of anti-cancer peptides, and output a feature matrix; the task-specific layer is ...
Embodiment 2
[0130] This embodiment provides a polypeptide anticancer function recognition system, which specifically includes the following modules:
[0131] A polypeptide sequence obtaining module, which is used to obtain a polypeptide sequence;
[0132]Anti-cancer function recognition module, which is used to input the polypeptide sequence into the trained polypeptide drug anti-cancer function predictor to obtain whether the polypeptide has anti-cancer function;
[0133] Wherein, the polypeptide drug anticancer function predictor includes an embedding layer, a coding layer and a task-specific layer, and the embedding layer is used to convert each residue of the received polypeptide sequence into a low-dimensional dense vector and output it in the form of a matrix; The encoding layer is used to capture the context of each residual embedding vector at different positions from the matrix output by the embedding layer, and learn the discriminative features of anticancer peptides, and output...
Embodiment 3
[0136] This embodiment provides a computer-readable storage medium, on which a computer program is stored. When the program is executed by a processor, the steps in the above-mentioned polypeptide anti-cancer function recognition system are realized.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com