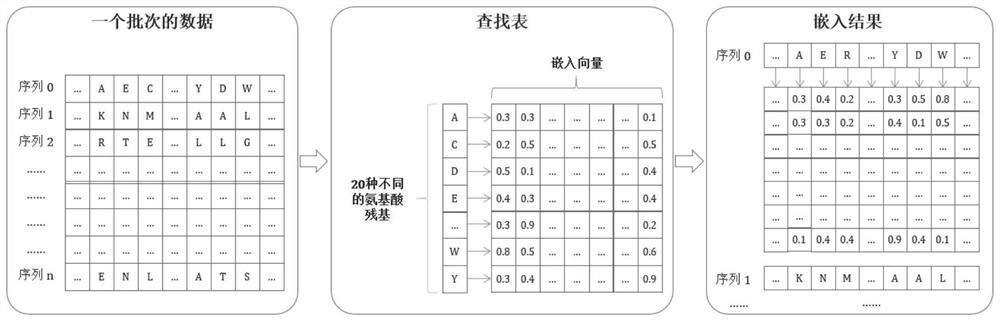
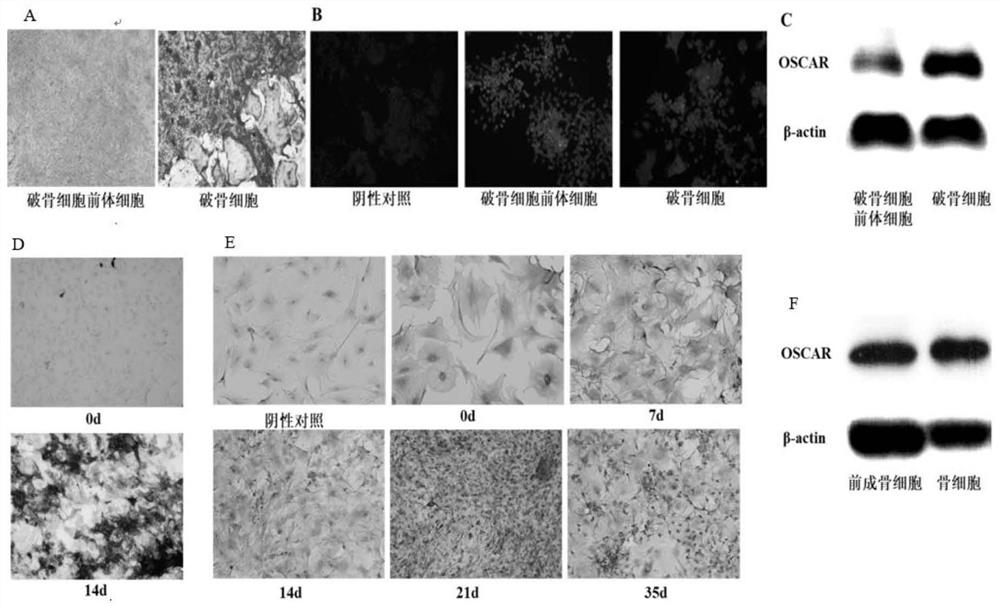
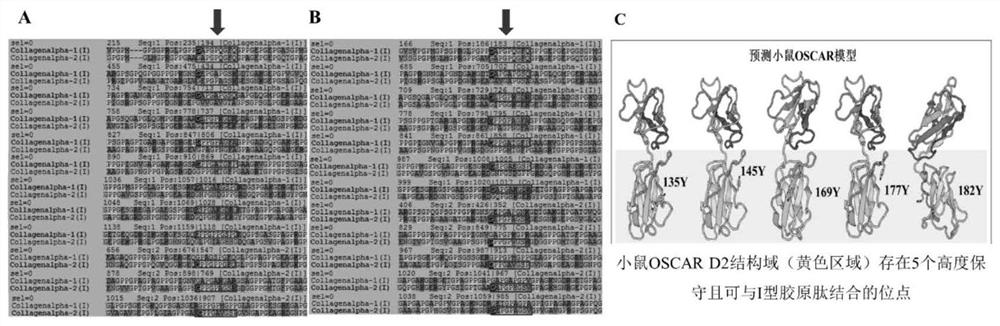
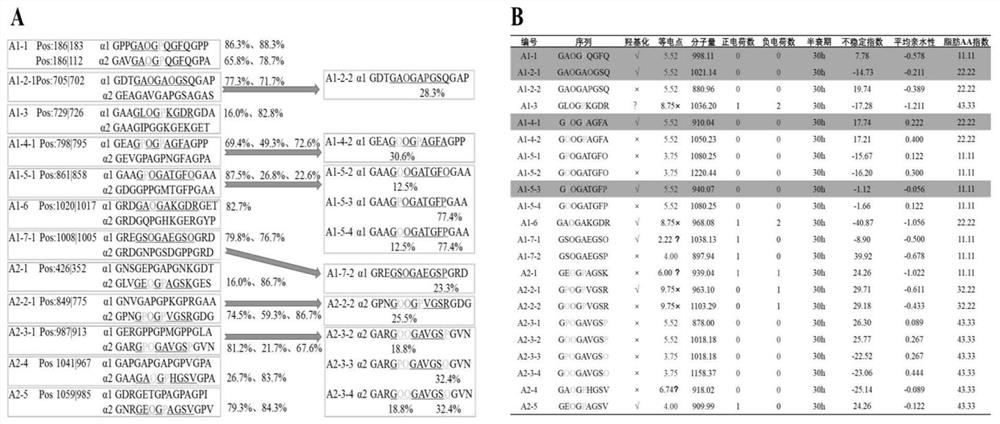
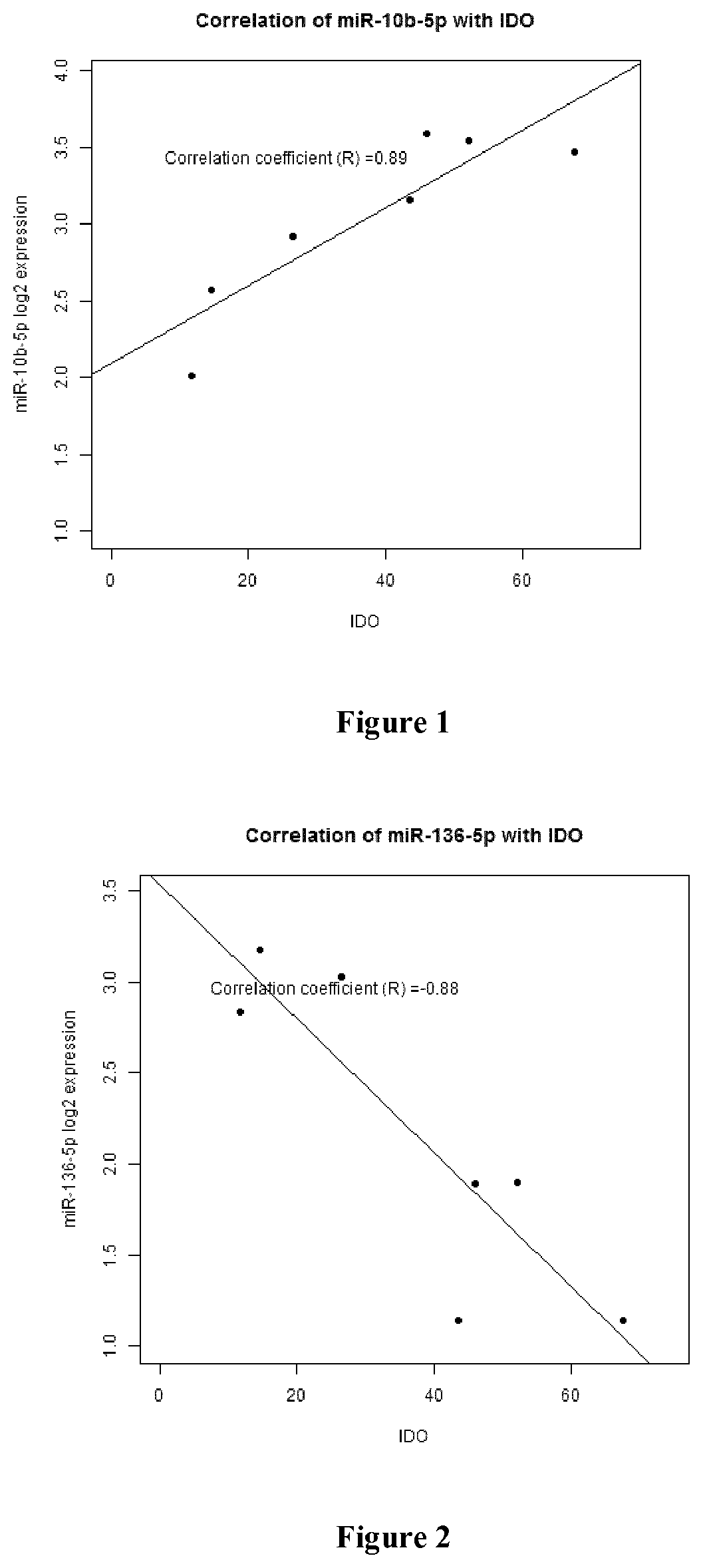
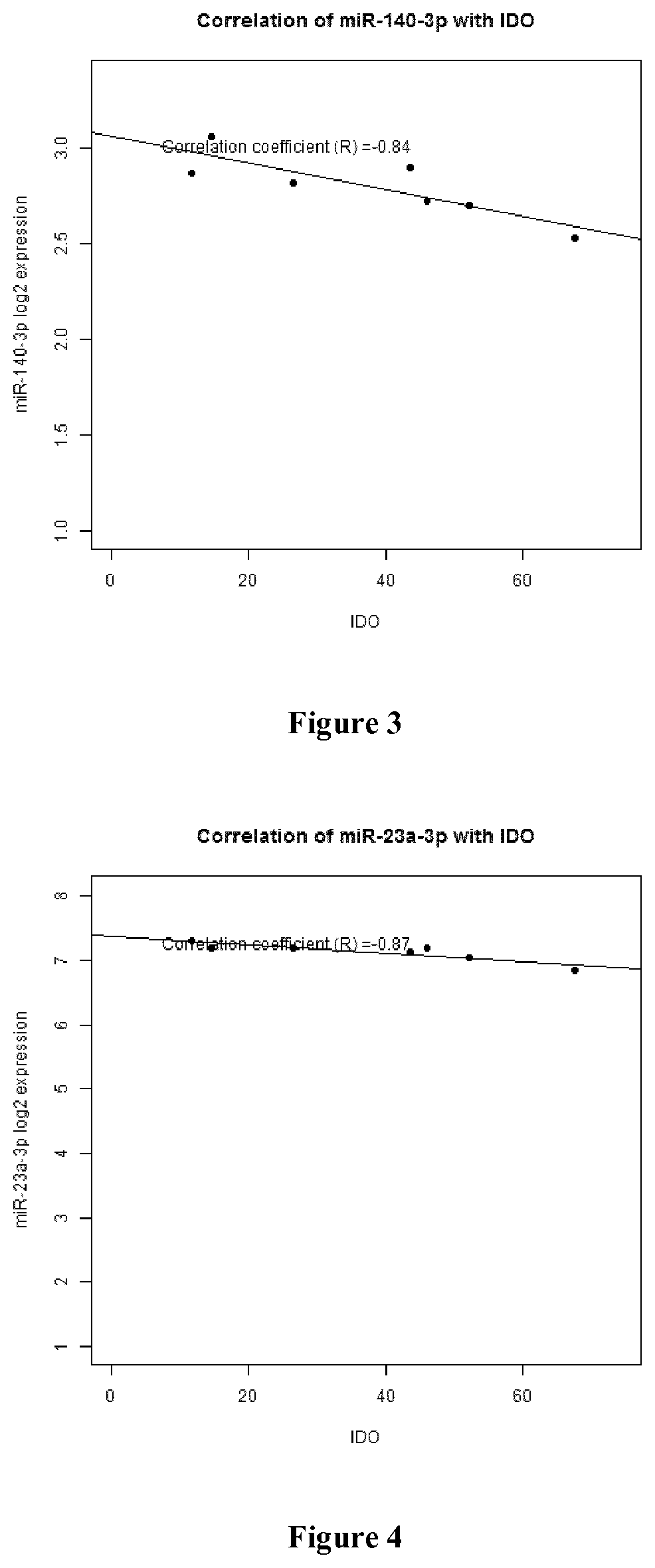
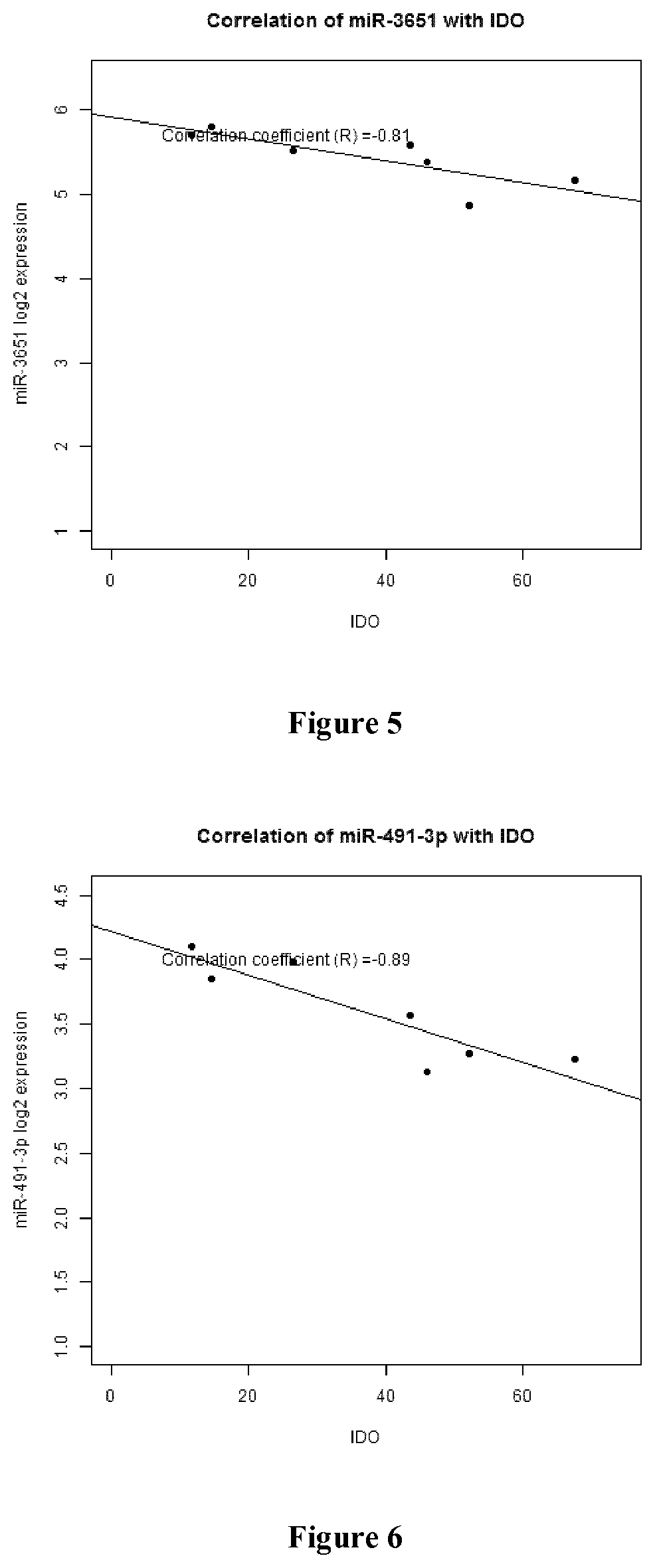
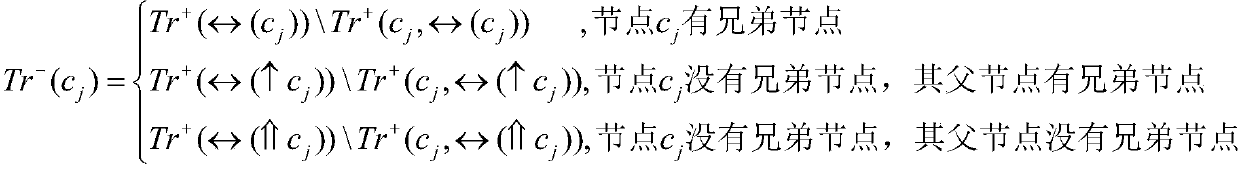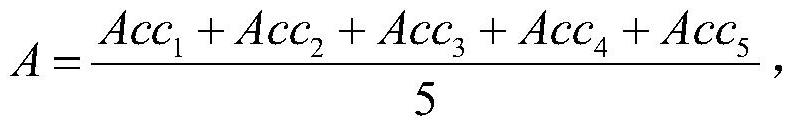Patents
Literature
35 results about "Functional prediction" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method for screening HRDs disease-causing mutation and gene chip hybridization probe designing method involved in same
ActiveCN103667438AClear relationshipScreening benefits are highMicrobiological testing/measurementDNA preparationDiseaseHybridization probe
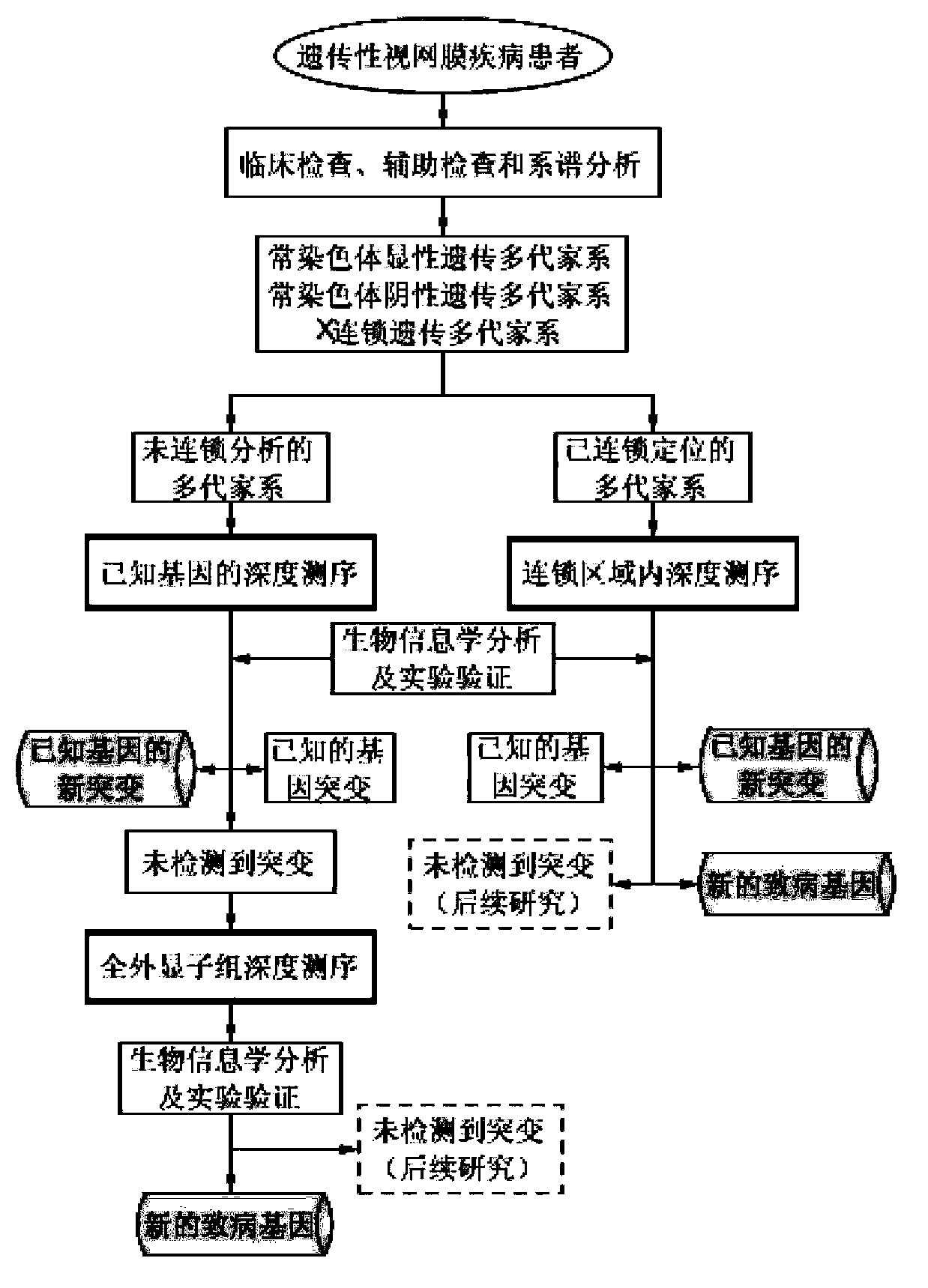
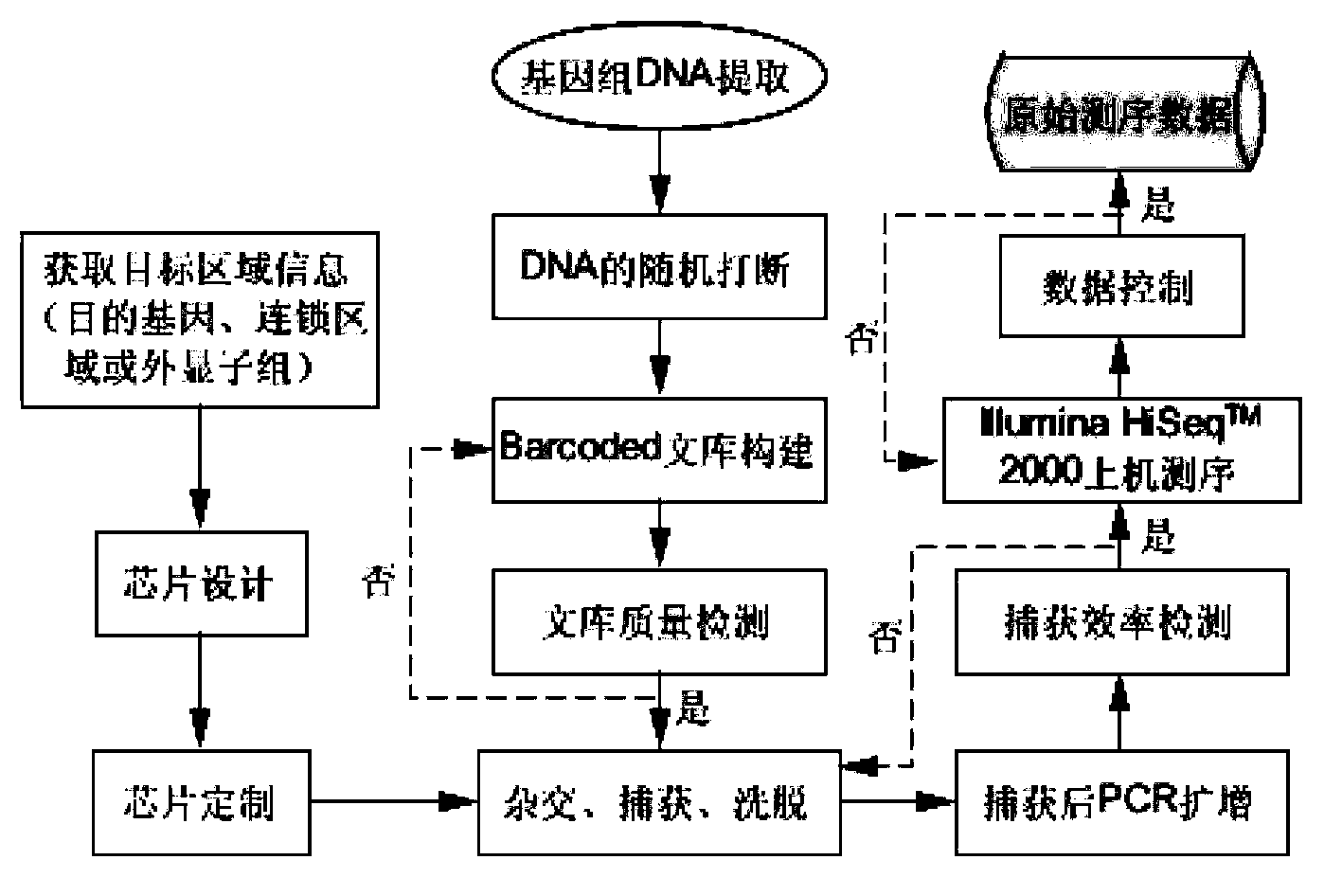
The invention belongs to the field of biological medicines, and relates to a method for screening HRDs disease-causing mutation and a gene chip hybridization probe designing method involved in the same. The method for screening the HRDs disease-causing mutation comprises the steps of (1) establishing an HRDs genetic resource repository; (2) designing and synthesizing a gene chip hybridization probe of an HRDs disease-causing gene, and integrating the gene chip hybridization probe onto a gene chip; (3) capturing a target area by utilizing the prepared gene chip and executing the depth sequencing; (4) analyzing the sequencing data on the aspect of bioinformatics, and screening the candidate disease-causing gene; (5) functionally predicting a newly-discovered splicing gene mutation site. By establishing the high-efficient HRDs target gene capturing technology, adopting the depth sequencing as a means and confirming the efficiency of the HRDs capturing chip, a high-efficient credible biological information analysis model is established.
Owner:赵晨
Predicting software product quality
ActiveUS9619363B1Reliability/availability analysisSoftware testing/debuggingParallel computingFunctional prediction
Owner:MAPLEBEAR INC
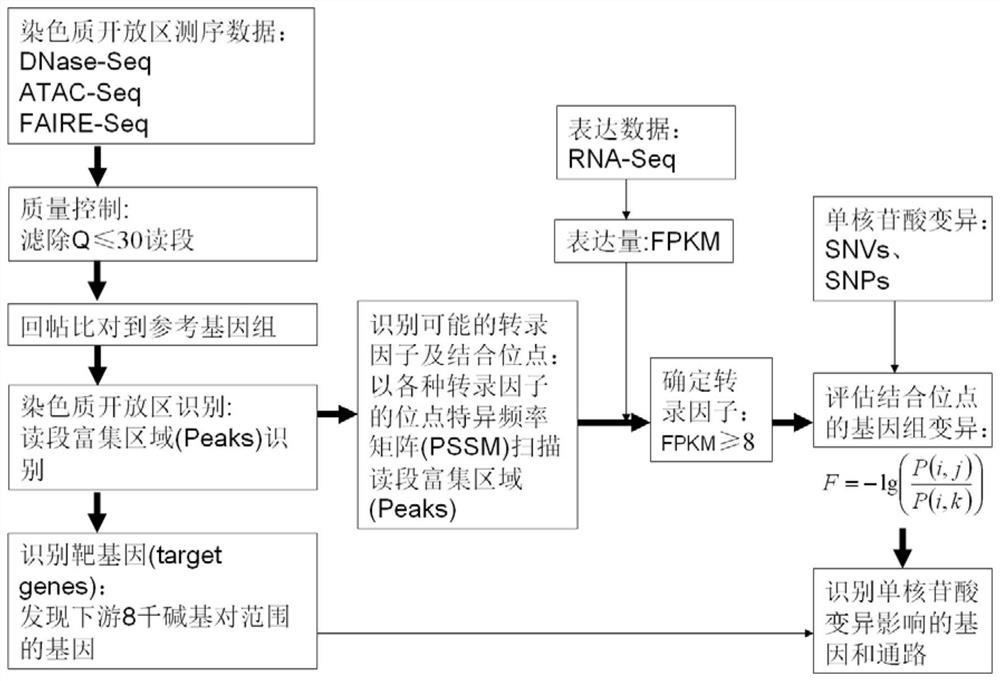
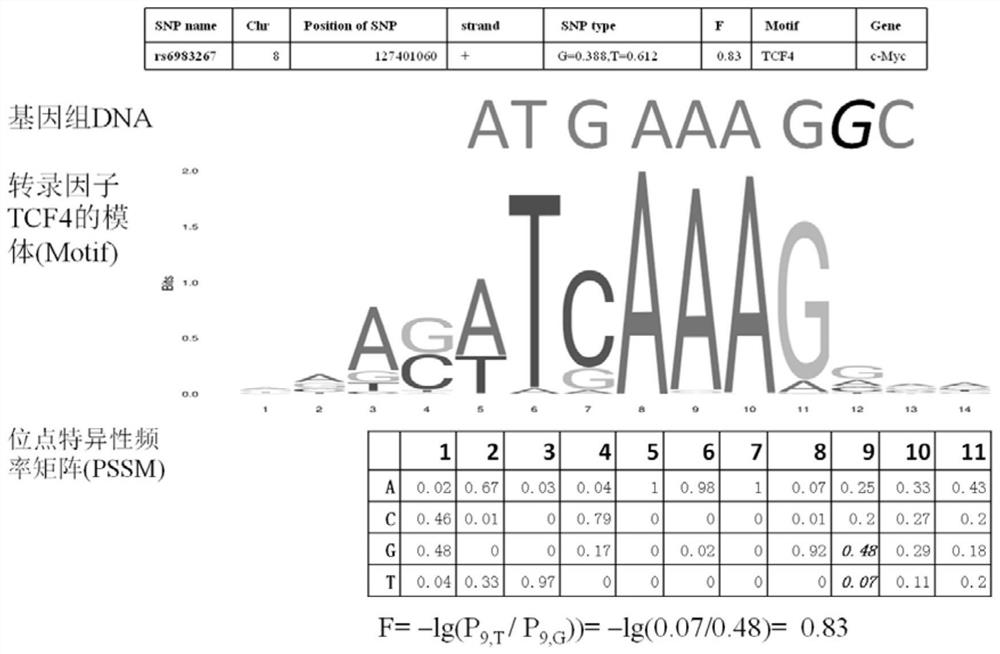
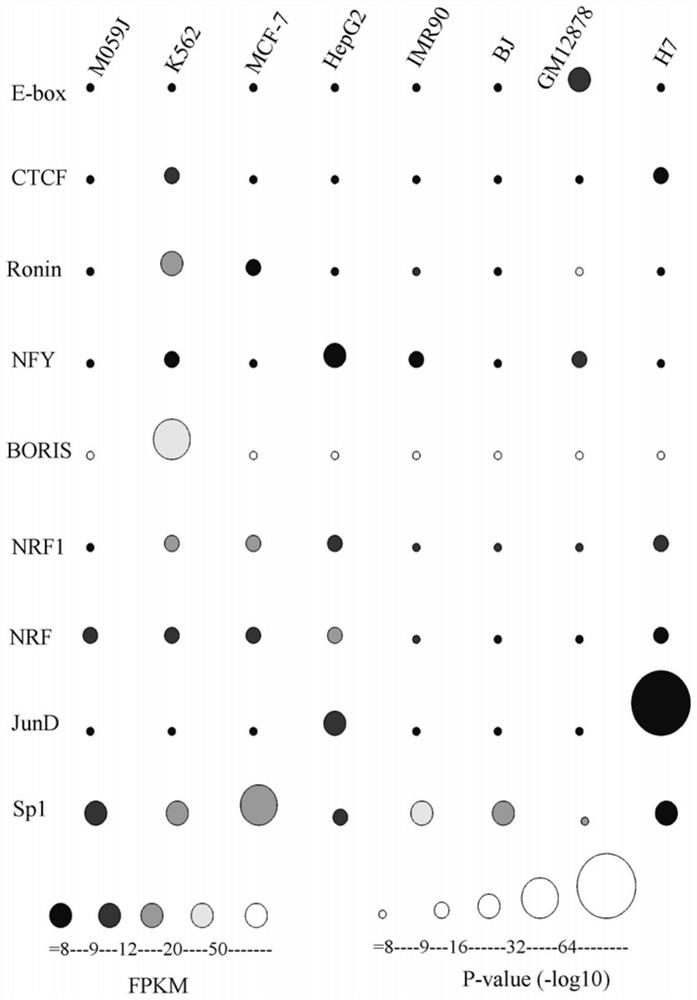
A method for functional prediction of single nucleotide genomic variation in a non-coding region
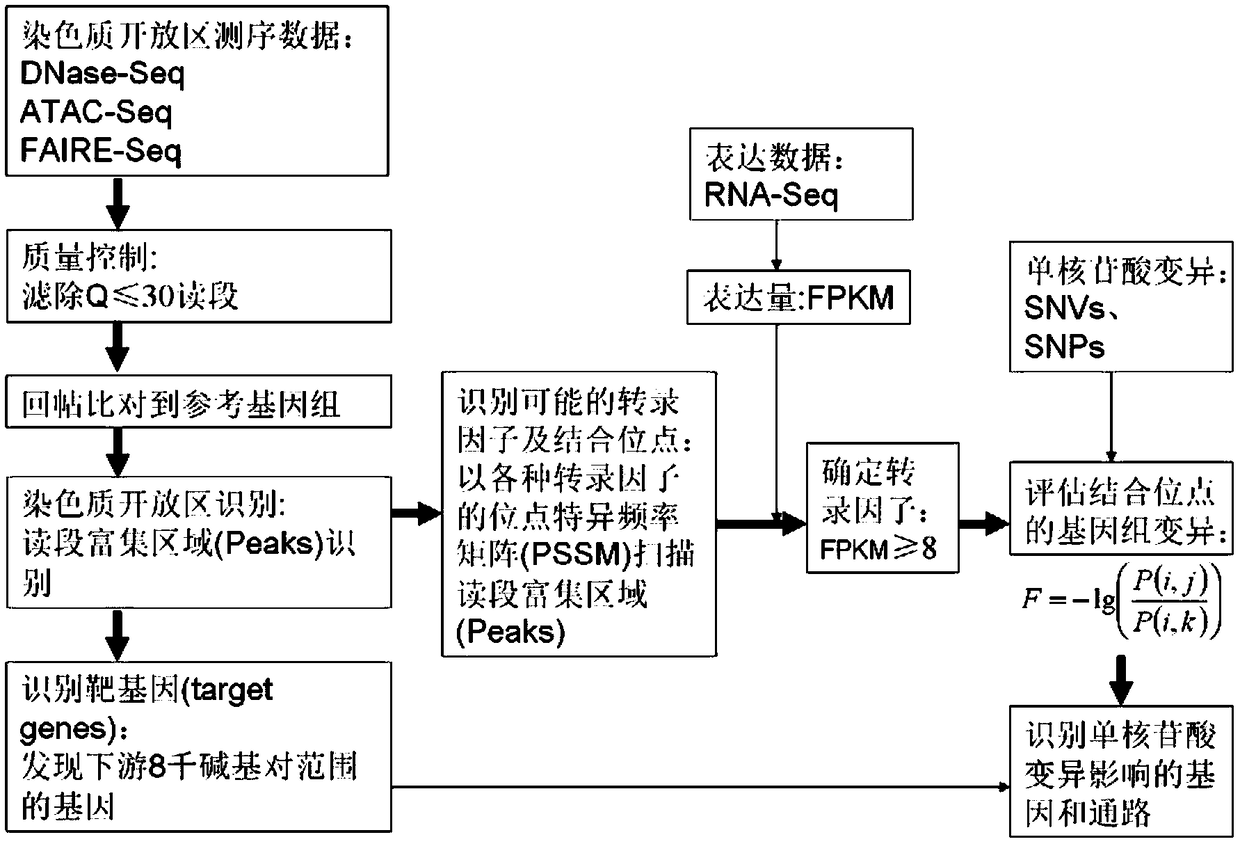
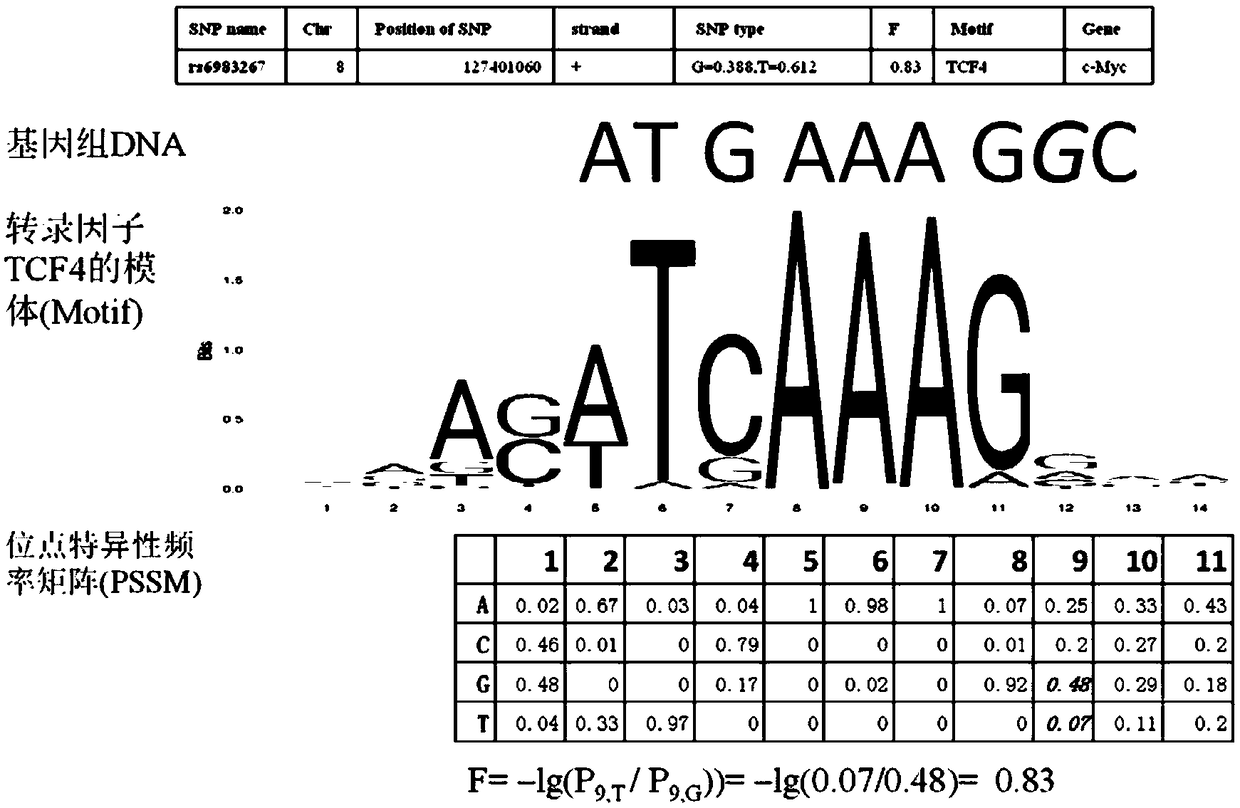
ActiveCN109033751ALow sequencing depth requirementAvoid time costSpecial data processing applicationsGenomic mutationNucleotide
The invention discloses a method for functional prediction of single nucleotide genomic variation in a non-coding region, which comprises the following steps: 1) identifying a chromatin open region; 2) identifying transcription factor bin sites; 3) assessing the role of single nucleotide variations: Based on the site-specific frequency matrix of transcription factors, the effect of single nucleotide variations in the transcription factor binding site region on transcription factor binding is calculated, and the single nucleotide variations that significantly change the binding capacity of transcription factors are identified; the role of single nucleotide variations is further evaluated by viewing the biological pathway of the target gene of the transcription factor. The method recognizesa variety of transcription factors and their binding sites at one time through chromatin open region information and gene expression information, and realizes the functional annotation of non-coding region genomic mutation.
Owner:SOUTHEAST UNIV
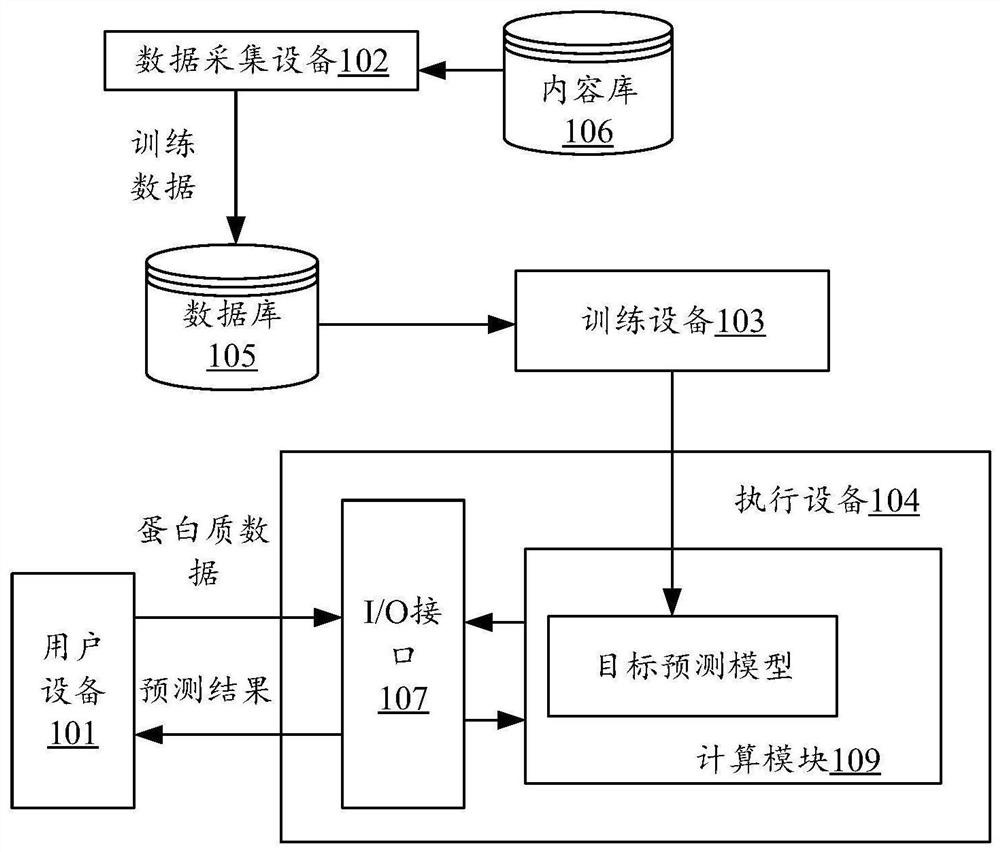
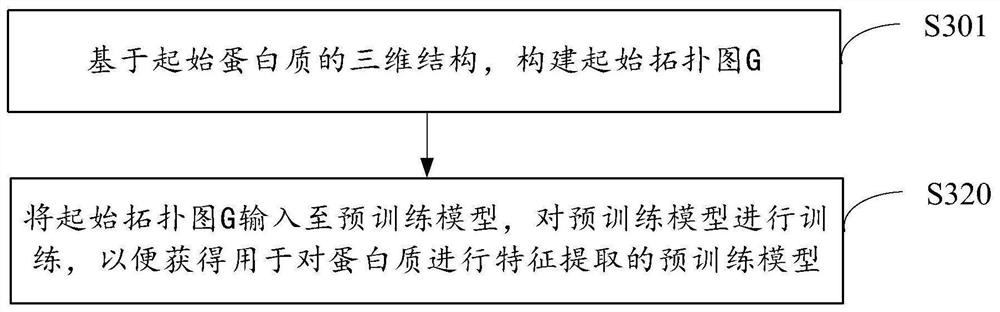
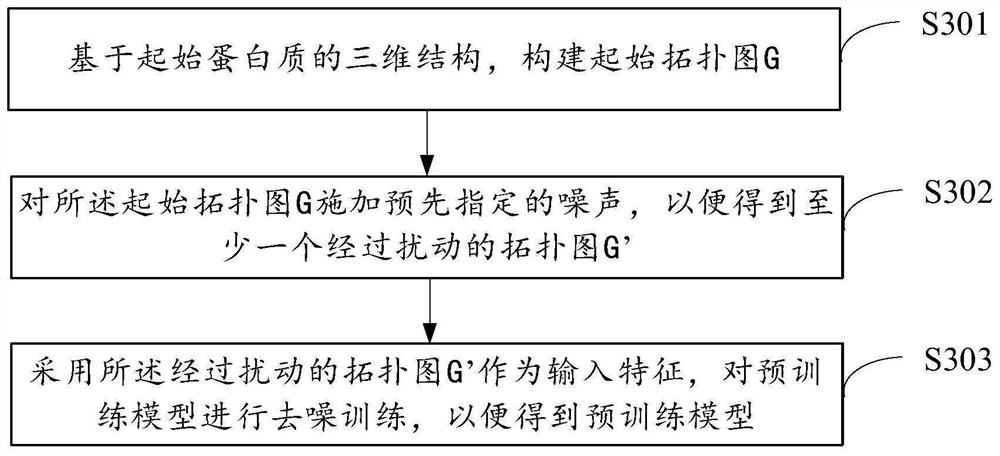
Method and device for model training, protein feature extraction and function prediction
PendingCN114333980AEfficient extractionImprove effectivenessBiostatisticsNeural architecturesFeature extractionAlgorithm
The embodiment of the invention provides a model training, protein feature extraction and function prediction method and device.The model training method comprises the steps that based on a three-dimensional structure of initial protein, an initial topological graph G is constructed, the initial topological graph G adopts amino acid residues of the initial protein as nodes, and the initial topological graph G adopts the amino acid residues of the initial protein as the nodes; the initial topological graph G adopts adjacent amino acid residue pairs as edges; and inputting the initial topological graph G into a pre-training model, training the pre-training model so as to obtain the pre-training model for performing feature extraction on the protein, the pre-training model comprising a graph neural network with SE (3) and other degeneration. Therefore, the prediction precision of related prediction such as protein feature extraction can be improved, the working cost is reduced, and the prediction efficiency is improved.
Owner:TENCENT TECH (SHENZHEN) CO LTD
Data processing apparatus, method, and computer program product for user objective prediction
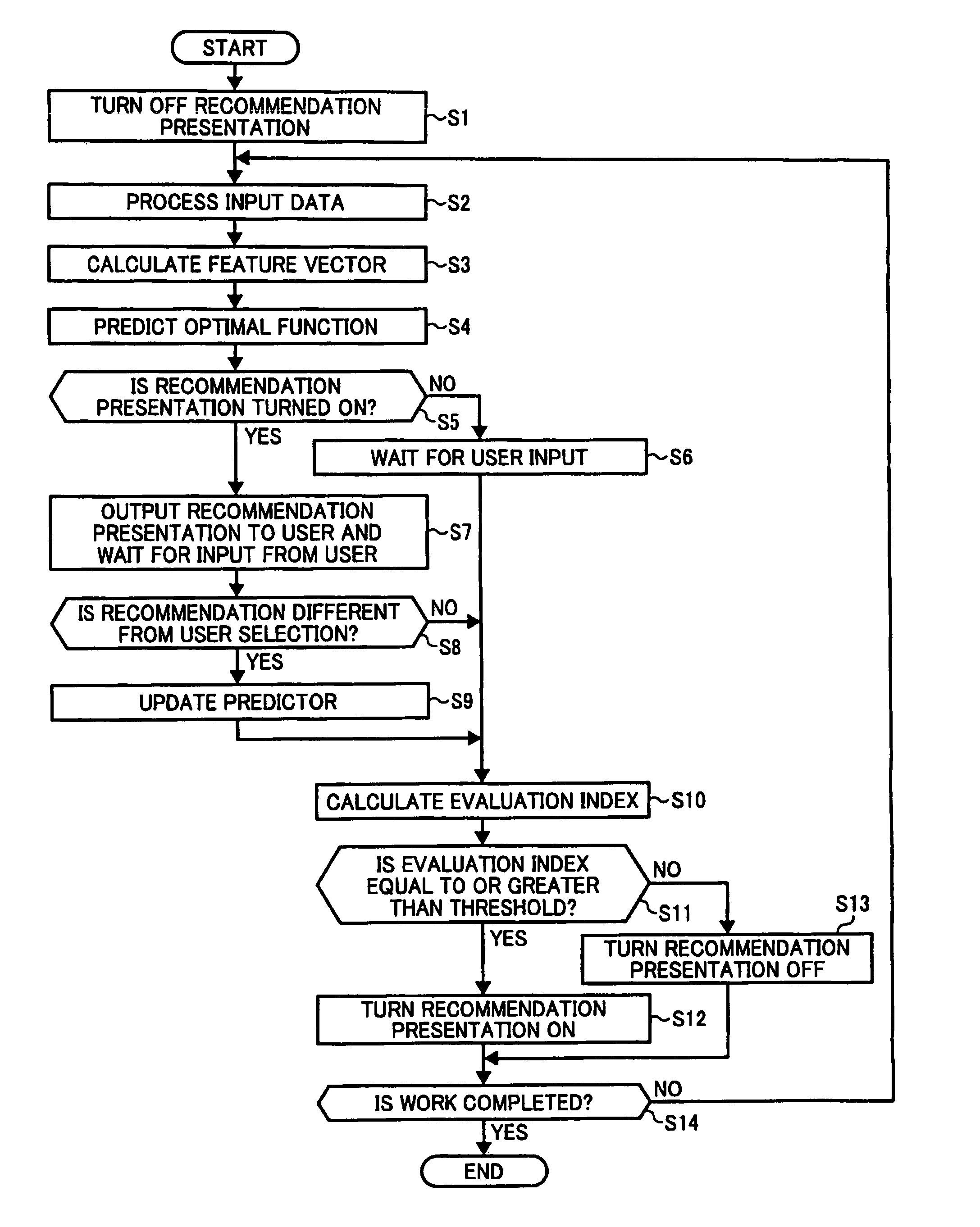
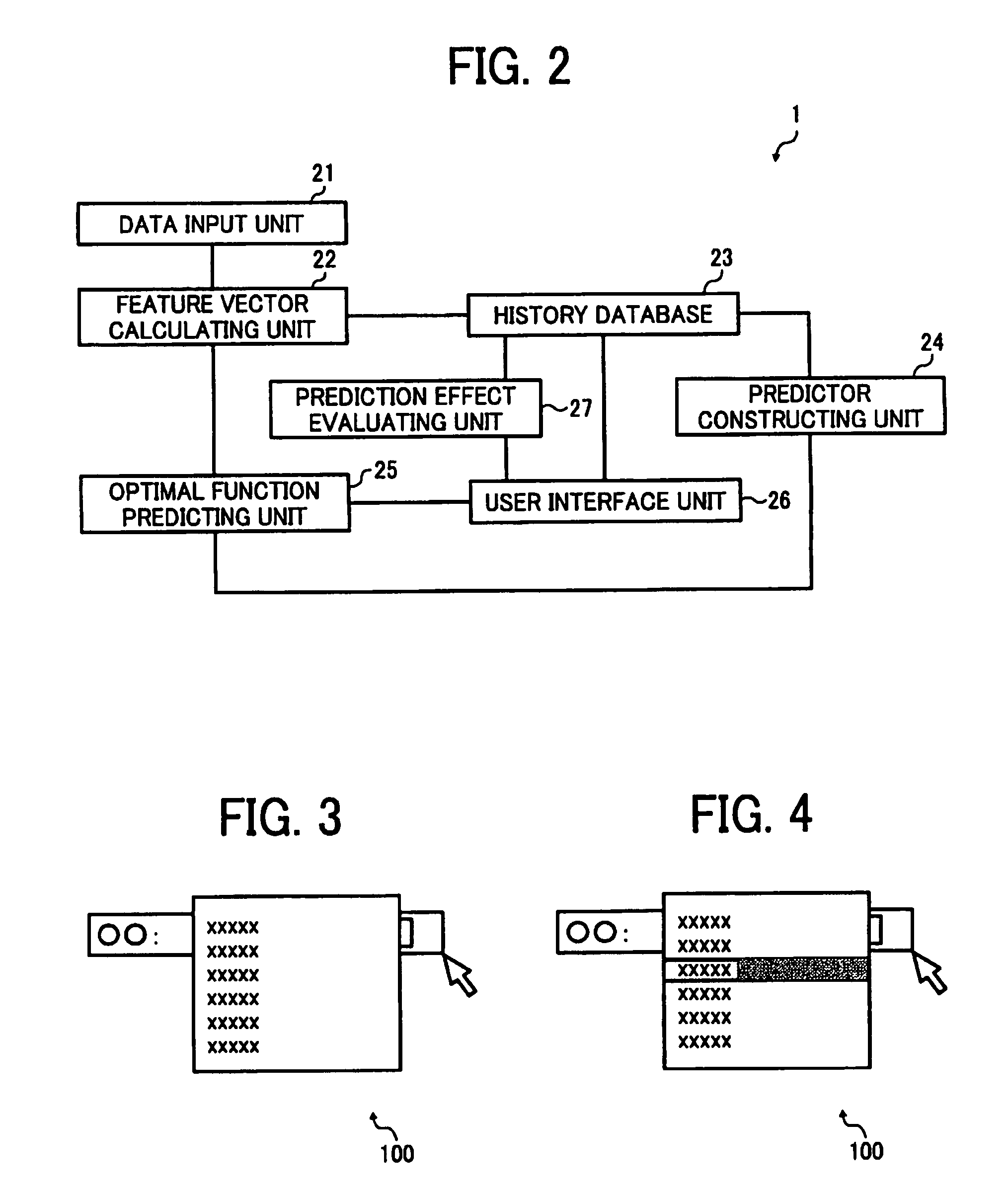
InactiveUS8321367B2Character and pattern recognitionOffice automationFeature vectorRecognition algorithm
A data processing apparatus includes an input unit that receives input data, a calculating unit that calculates a feature vector, a selection receiving unit that presents processing options to a user and receives a user selection, a storing unit stores processing history information, and a predictor constructing unit that constructs a predictor for an identifying algorithm that predicts optimal processing for input data based on processing history information stored in a history database. An optimal function predicting unit predicts optimal processing based on a feature vector calculated for new data by using the predictor and an evaluating unit evaluates work efficiency for the predicted optimal processing, wherein the evaluating unit is configured to evaluate work efficiency for the optimal processing based on a prediction accuracy of the predictor and a user specific working time that is required for each user to select one of the processing options with the selection receiving unit. Finally, a presenting unit presents the optimal processing to a user when an evaluation result obtained by the evaluating unit is equal to or greater than a threshold.
Owner:RICOH KK
Screening method for tobacco antiviral regulatory genes and application of screening method
InactiveCN109610009AEasy accessSimple and fast operationMicrobiological testing/measurementVector-based foreign material introductionSequence analysisCDNA library
The invention provides a screening method for tobacco antiviral regulatory genes and an application of the screening method. The screening method comprises the following steps: firstly constructing atobacco cDNA library by using a virus-induced gene silencing (VIGS) vector as a target vector, and performing homogenization treatment to obtain a VIGS-cDNA library capable of being directly used forgene silencing screening analysis; transforming VIGS-cDNA library plasmid into agrobacteria to obtain an agrobacterium library; performing spread plate culture on the agrobacterium library to obtain single colonies, performing VIGS silencing analysis on each single colony, and performing screening to obtain antiviral-related colonies; and performing sequencing analysis on genes corresponding to the screened antiviral-related colonies, performing homology analysis on the obtained sequences, and performing function prediction to obtain the antiviral regulatory genes. The method provided by the invention has the advantages of simple operation, no need to construct transgenic plants or mutants, short time consuming, comprehensive screening and high efficiency, is particularly suitable for screening of mutant lethal genes, and has broad application prospects.
Owner:GUIZHOU TOBACCO SCI RES INST
Analysis method for succession of microbial communities during tobacco mellowing
InactiveCN111187850AImprove qualityReduce fermentation costsTobacco treatmentMicrobiological testing/measurementBiotechnologyNicotiana tabacum
The invention relates to an analysis method for succession of microbial communities during tobacco mellowing, and belongs to the technical field of environmental microbes. In order to define microbialpopulation structures and dynamic changes thereof and predict microbial gene functions during tobacco mellowing, the invention provides the analysis method for the succession of the microbial communities during tobacco mellowing, and the analysis method comprises the following steps of (1) performing sampling: taking naturally mellowed tobacco samples at the beginning of mellowing at intervals ofthe same time; (2) constructing gene libraries: extracting genomic DNA of the samples collected in each time period, and constructing a 16S ribosomal RNA (16S rRNA) gene library and an internal transcribed spacer region gene library; and (3) analyzing data of each gene library constructed in the step (2) to obtain the rule of the succession of the microbial communities during tobacco mellowing. The analysis method can be used for giving guidance on artificially mellowing tobacco and establishing the microbial communities.
Owner:QINGDAO INST OF BIOENERGY & BIOPROCESS TECH CHINESE ACADEMY OF SCI
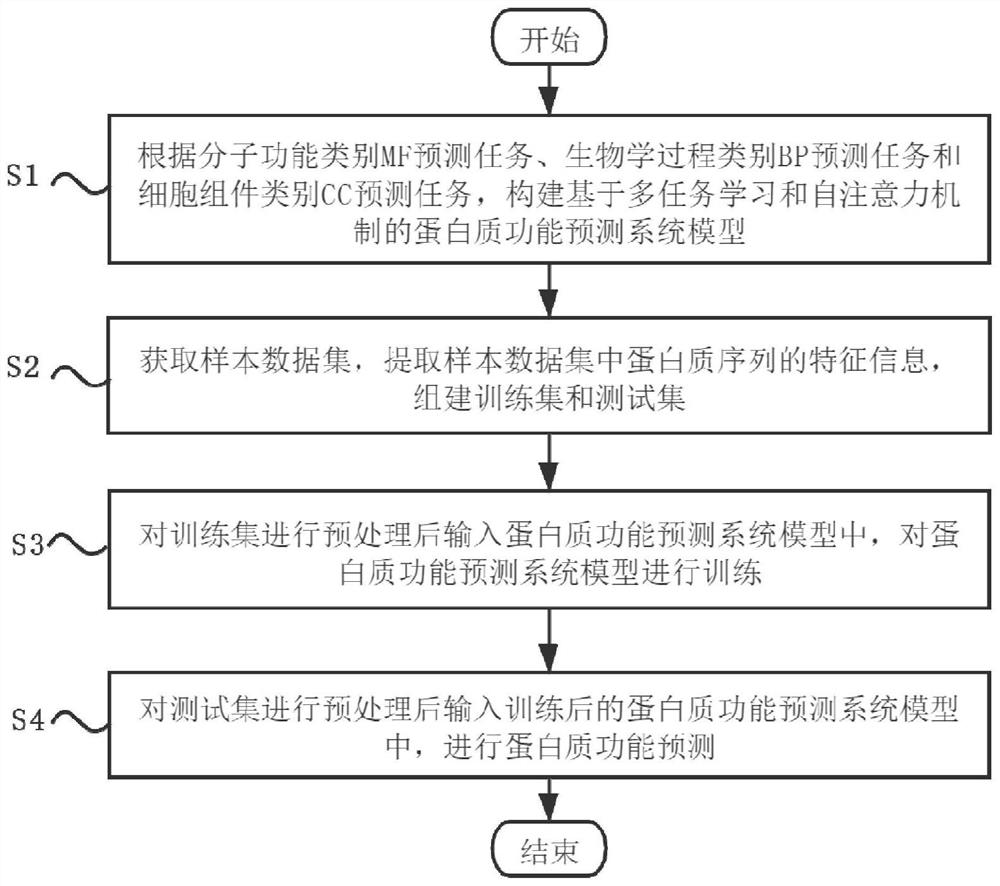
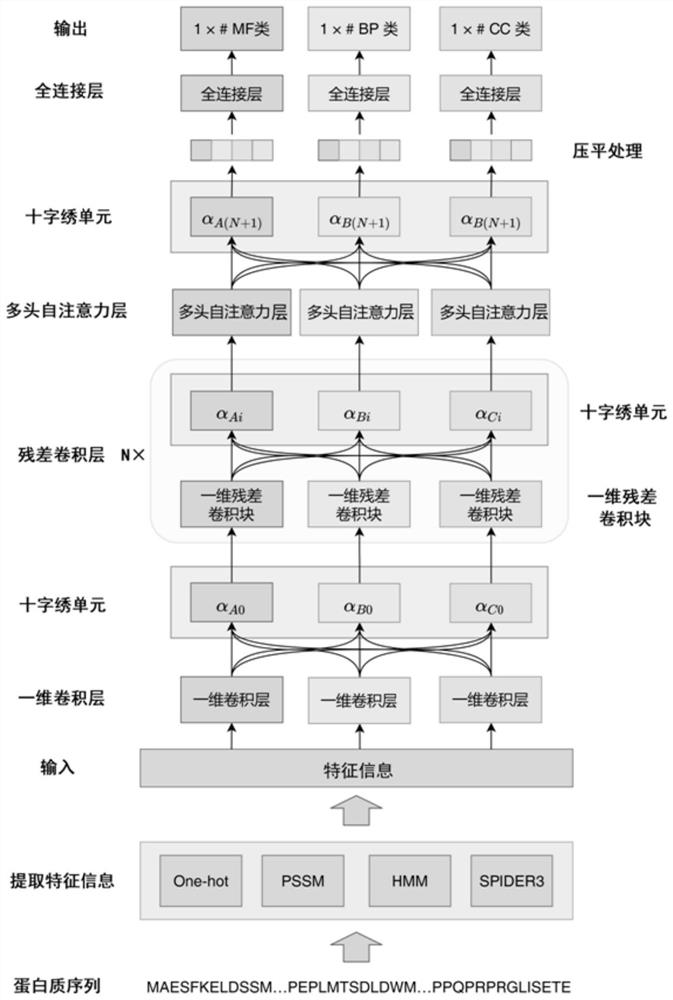
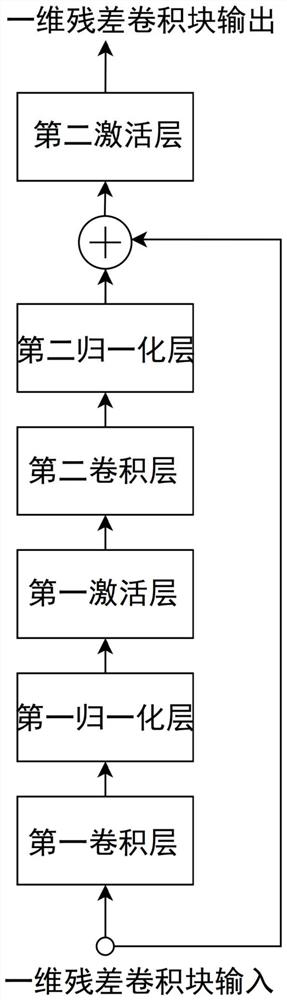
Protein function prediction method combining multi-task learning and self-attention mechanism
The invention discloses a protein function prediction method combining multi-task learning and a self-attention mechanism, and the method comprises the following steps: predicting a task according toa molecular function type MF, a biological process type BP, and a cell module type CC; constructing a protein function prediction system model based on multi-task learning and a self-attention mechanism; obtaining a sample data set, extracting feature information of protein sequences in the sample data set, and establishing a training set and a test set; preprocessing the training set, inputting the preprocessed training set into a protein function prediction system model, and training the protein function prediction system model; and preprocessing the test set, inputting the preprocessed testset into the trained protein function prediction system model, and carrying out protein function prediction. According to the method, prediction of three ontology theories is regarded as three prediction tasks, prediction is performed by establishing a protein function prediction system model based on multi-task learning and a self-attention mechanism, and the accuracy of protein function prediction is improved.
Owner:SUN YAT SEN UNIV
CircRNA function prediction method based on cascade decision system
PendingCN111755070AStability improvements improvedImprove stabilityProteomicsGenomicsFeature extractionDecision system
In order to overcome the defects in the prior art, the invention aims to predict the function of the circRNA by using the provided cascade decision system in combination with the multi-classificationmodel of the LightGBM method. The technical scheme adopted by the invention mainly comprises the following steps of: (1) inputting the CircRNA of a big data sample in a (. bed) file form; (2) mappingthe CircRNA (. bed) file according to related information such as a starting site and the like to obtain a CircRNA sequence information (. fasta) file; (3) proposing a feature extraction and fusion method, and extracting a CircRNA feature; (4) proposing an A-type judgment system, and performing function prediction on the coded circRNA; (5) predicting other CircRNA by utilizing the LightGBM (LightGBM) algorithm; (6) according to a multi-classification model of a lightGBM algorithm, carrying out sampling and feature sampling of sample data by using core algorithms GOSS and EFB, mapping continuous features into discrete buckets by using a Histogram-based algorithm, and discretizing continuous variables; and (7) obtaining the optimal parameters of the model by adjusting the maximum depth of the tree, the minimum record number of the leaves, the data proportion used in each iteration and other parameters.
Owner:SUN YAT SEN UNIV
Object functionality prediction method and device, computer equipment and storage medium
ActiveCN108921952AImprove versatilityImprove accuracyImage data processingThree-dimensional object recognitionFeature vectorComputer graphics (images)
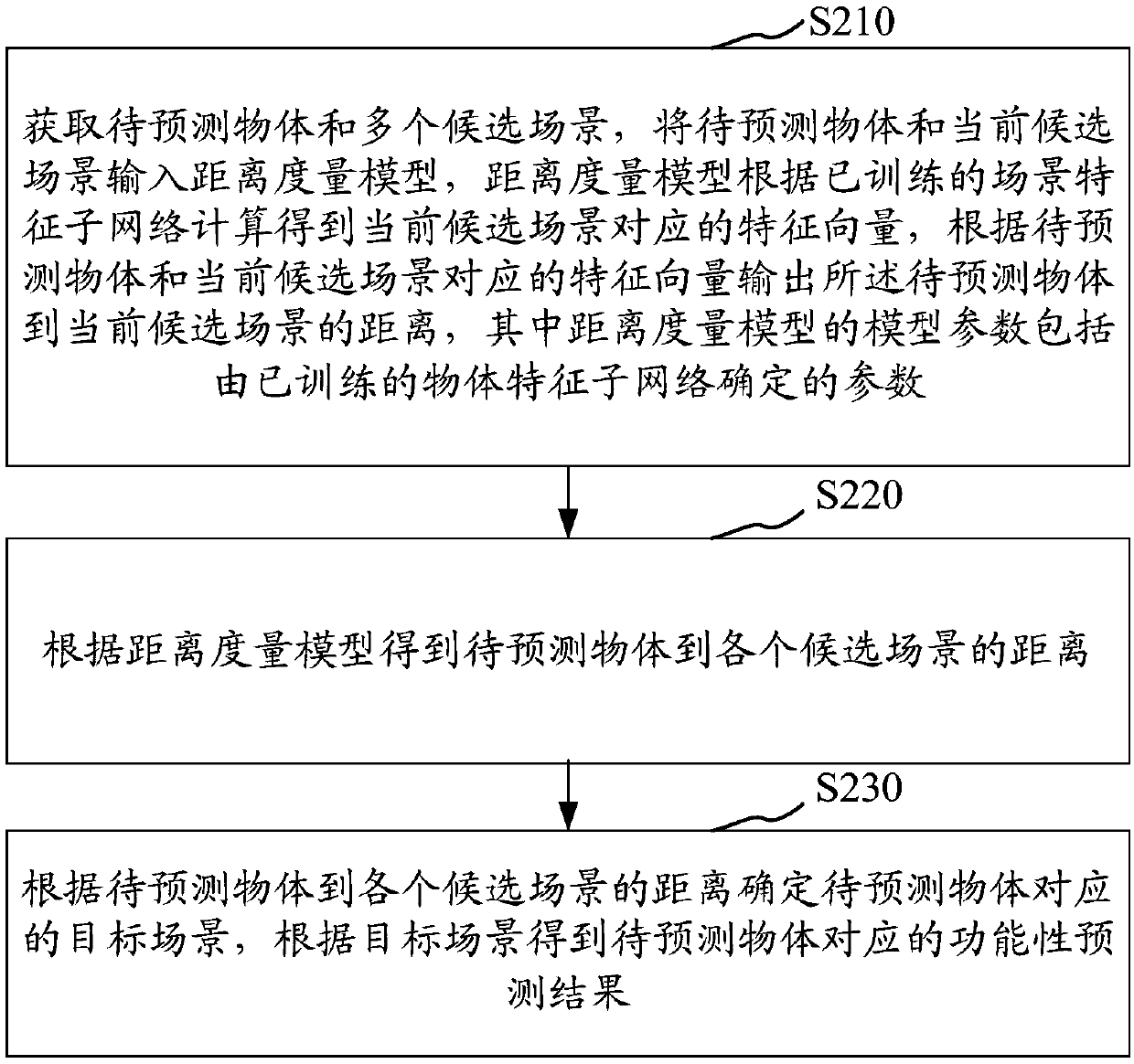
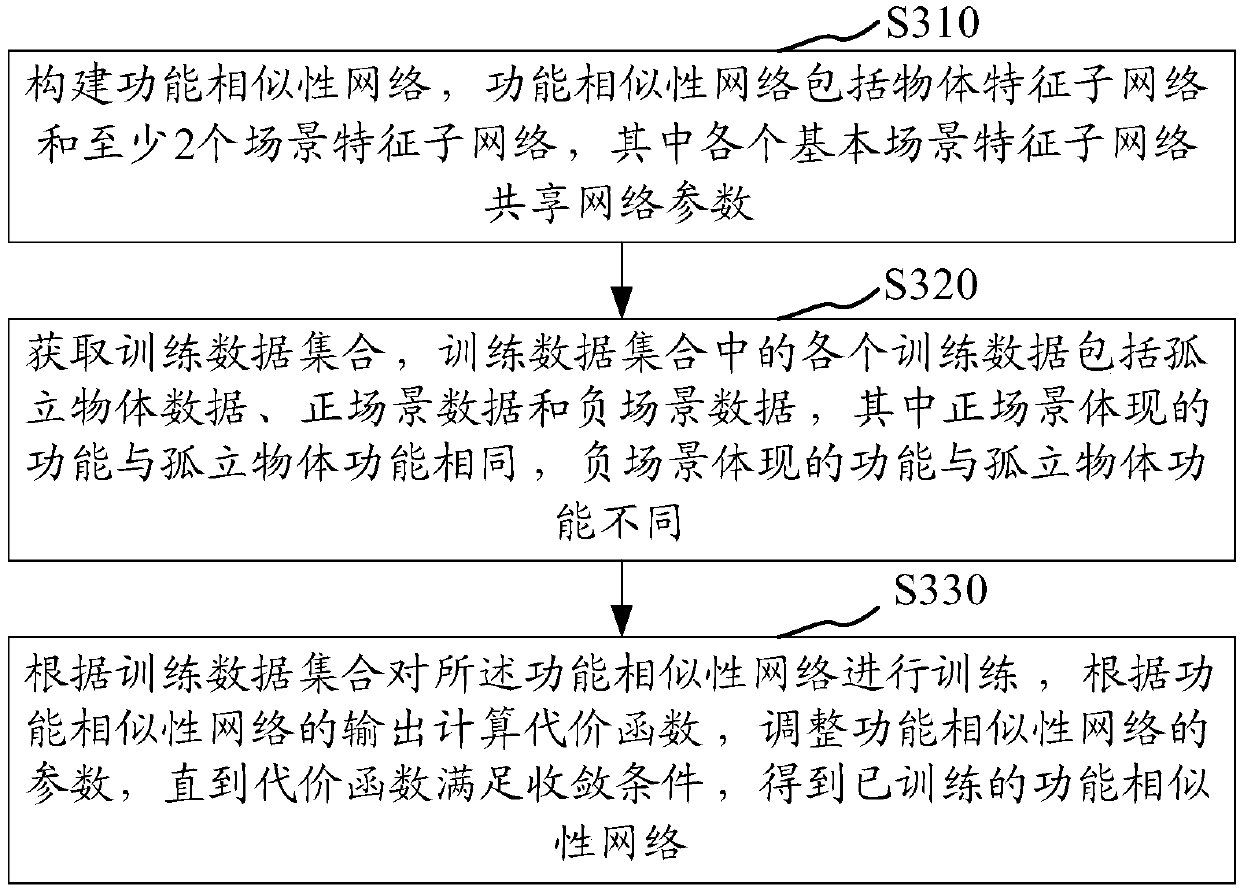
The invention relates to an object functionality prediction method and device, computer equipment and a storage medium. The method comprises the steps of acquiring a to-be-predicted object and a plurality of candidate scenes; inputting the to-be-predicted object and the current candidate scenes to a distance measurement model, performing calculation by the distance measurement model according to atrained scene feature sub-network to obtain eigenvectors corresponding to the current candidate scenes, and outputting distances from the to-be-predicted object to the current candidate scenes according to the to-be-predicted object and the eigenvectors corresponding to the current candidate scenes, wherein model parameters of the distance measurement model include parameters determined by a trained object feature sub-network; obtaining the distances from the to-be-predicted object to the candidate scenes according to the distance measurement model; determining a target scene corresponding tothe to-be-predicted object according to the distances from the to-be-predicted object to the candidate scenes; and obtaining a functionality prediction result corresponding to the to-be-predicted object according to the target scene. The object functionality prediction universality can be improved.
Owner:SHENZHEN UNIV
Polypeptide anticancer function identification method, system, medium and equipment
PendingCN113593632AImprove accuracyCharacter and pattern recognitionNeural architecturesPharmaceutical drugFunctional prediction
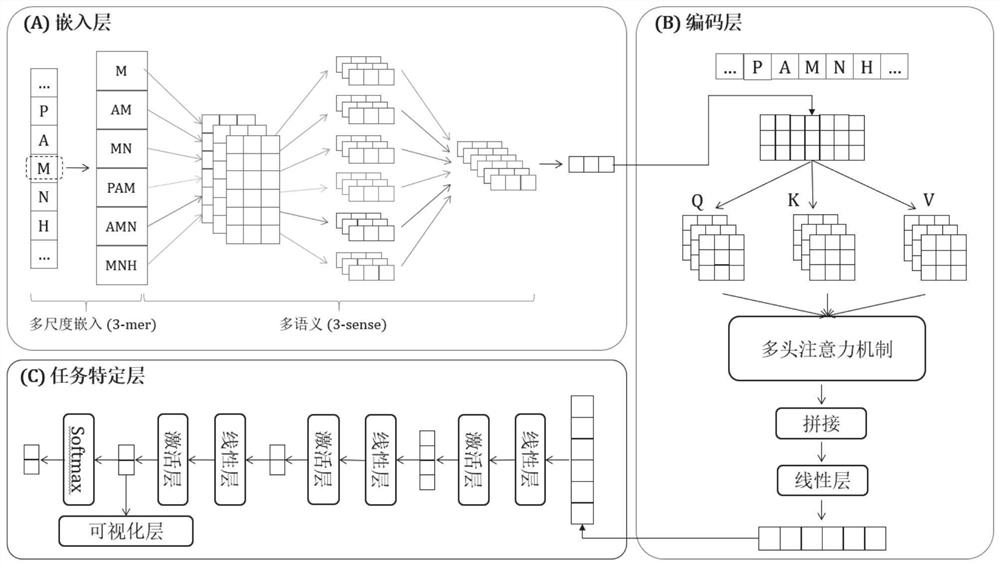
The invention belongs to the technical field of biological information, and provides a polypeptide anticancer function identification method, system, medium and equipment. The identification method comprises the following steps: acquiring a polypeptide sequence; inputting the polypeptide sequence into the trained anticancer function predictor of the polypeptide drug to obtain whether the polypeptide has an anticancer function or not; wherein the anticancer function predictor of the polypeptide drug comprises an embedding layer, a coding layer and a task specific layer, and the embedding layer is used for converting each residue of a received polypeptide sequence into a low-dimensional dense vector and outputting the low-dimensional dense vector in a matrix form; the coding layer being used for capturing contexts of each residual embedding vector at different positions from the matrix output by the embedding layer, learning identification characteristics of the anti-cancer peptide and outputting a characteristic matrix; and the task specific layer being used for judging the probability that the polypeptide sequence belongs to the anti-cancer peptide according to the characteristic matrix.
Owner:SHANDONG UNIV
Bone remodeling regulating polypeptides and application
PendingCN111647042AImprove biostabilityEasy to synthesizeConnective tissue peptidesPeptide/protein ingredientsMedicineEngineering
The invention provides bone remodeling regulating polypeptides and application. At the early stage, on the basis of first experiment findings, through sequence alignment, structural analysis, physicochemical properties and function prediction, a solid-phase polypeptide synthesis method is designed and utilized to newly synthesize the bone remodeling regulating polypeptides. The polypeptides have the characteristics of short length, simple synthesis, low cost, lower cytotoxicity, higher biological stability, moderate half-life period, good targeting ability and high bone absorption regulation capacity, differentiation and functions of osteoclasts and osteoblasts can be controlled by regulating the concentration of the synthesized bone remodeling regulating polypeptides to realize ordered regulation of bone absorption and bone formation, and the polypeptides have broad potential application prospects.
Owner:THE SECOND XIANGYA HOSPITAL OF CENT SOUTH UNIV
Drug target prediction method for keeping consistency of chemical properties and functions of drug and system thereof
PendingCN113345535AMaintain chemical similarityGuaranteed chemical propertiesMolecular designDiseasePharmaceutical drug
The invention provides a drug target prediction method for keeping consistency of chemical properties and functions of a drug and a system thereof, and belongs to the technical field of computer-aided drug research and development, and the method comprises the steps: obtaining a chemical fingerprint of a to-be-predicted drug; using the trained feature selection model to process the chemical fingerprint of the drug to obtain an interaction scoring matrix of the drug and the target; and taking the corresponding target with the highest score as the action target of the drug based on the interaction score matrix of the drug and the target. According to the invention, chemical properties and clinical functions of drugs are considered at the same time, and possible targets of the drugs are predicted; the feature vector of the drug is projected to a protein space and then projected to a disease space, a drug-target interaction prediction task is changed into a multi-label task from a traditional single-label classification task, and a complex repulsion relationship between the drug and protein is considered; by keeping the consistency of the chemical similarity and the functional similarity of the medicine, the consistency of the chemical property, the molecular mechanism and the clinical function of the medicine is kept.
Owner:NANKAI UNIV
Gene function prediction method based on r-svm and tpr rules
ActiveCN106126973BAchieve forecastSolve the multi-label problemCharacter and pattern recognitionProteomicsPositive sampleSvm classifier
A method for predicting gene function based on R-SVM and TPR rules relates to a method for predicting gene function. The invention can realize the prediction of the gene function, and can solve the multi-label problem and the hierarchical constraint problem existing when the classification algorithm is used to realize the prediction of the gene function. The present invention firstly uses genes with known functions as training samples to form a training set; for each node in the GO annotation scheme, a positive sample set and a negative sample set are constructed; for each node in the GO annotation scheme, select the node The attribute that contributes a lot to the classification of the function; through training, a set of R-SVM classifiers is obtained and the unknown samples are classified and predicted, and a set of preliminary R-SVM classification results are obtained; the classification results are converted into posterior probability values, using A weighted TPR ensemble algorithm for directed acyclic graph hierarchy to realize gene function prediction. The invention is applicable to the prediction of gene function.
Owner:黑龙江鉴成医学检验实验室有限公司
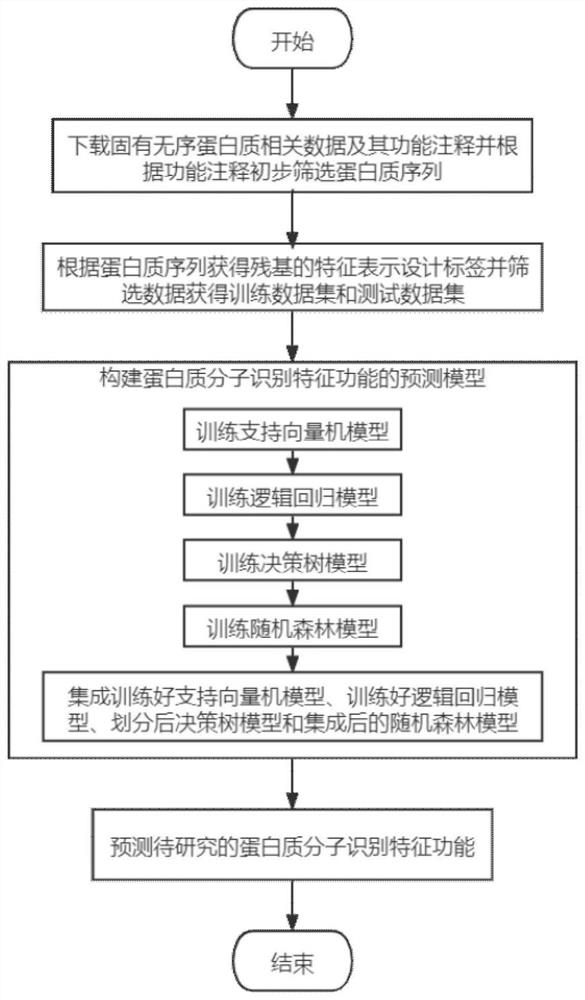
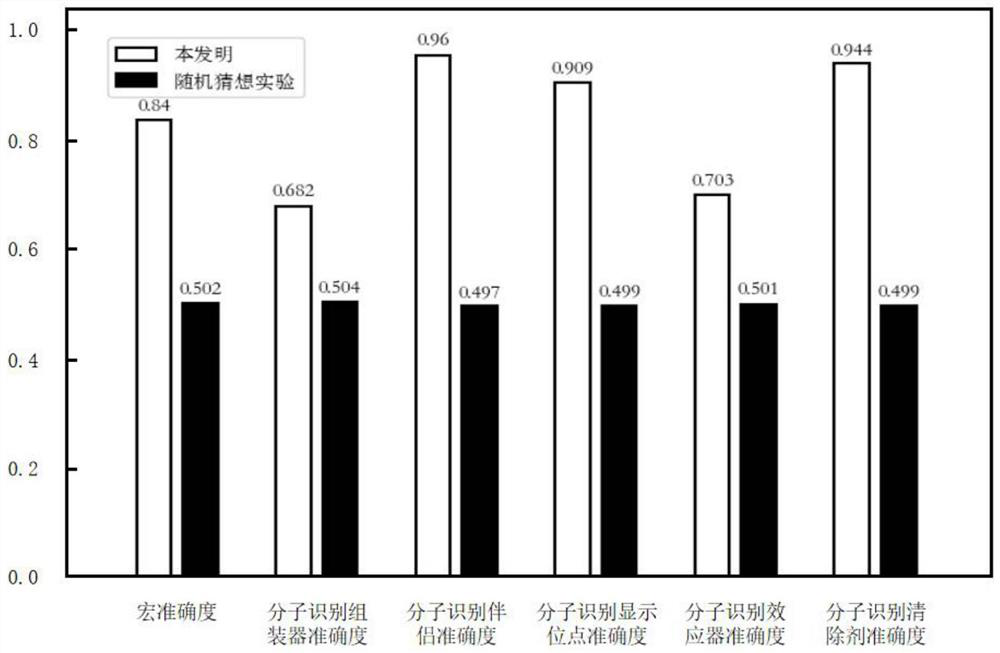
Molecular recognition feature function prediction method based on ensemble learning
PendingCN113936748AImprove accuracySimplify the experimental processMolecular entity identificationKernel methodsDrug targetEnsemble learning
The invention discloses a molecular recognition feature function prediction method based on ensemble learning, and mainly solves the problem that an existing molecular recognition feature predictor cannot further divide molecular recognition feature functions. According to the scheme, the method comprises the following steps: downloading inherent disordered protein data and functional annotations thereof, dividing training data and test data, performing feature representation on a protein sequence, and designing a residue tag of the protein sequence; selecting a single-input binary association strategy machine learning model; training different machine learning models by using the training data; integrating training results of different machine learning models by using an integration strategy to collect a prediction model; and inputting to-be-researched protein sequence data into the prediction model, and outputting a molecular recognition feature function prediction result of the protein. The method is simple in experimental process, low in resource consumption, low in cost and high in reliability of prediction results, can be used for predicting molecular recognition features in protein sequences, and provides reference for drug target acting positions.
Owner:XIDIAN UNIV
Functional prediction of cellular functions by means of microRNA expression profiling in mesenchymal stem cells
ActiveUS11266687B2Microbiological testing/measurementDigestive systemMesenchymal stem cellFunctional prediction
Non-coding RNA, such as miRNA, expression data derived from a cell population is used to infer the propensity of that cell population for a cellular functional effect for a pre-determined purpose, which effect is temporally, procedurally or interventionally separated from the cell population from which the expression data is derived. Thereby, the cellular functional effect of a cell population can be predicted in order to improve decisions and selections to be made relating to the use of cells, e.g. from cells deriving from different donors or batches, for use in bioprocess application or cell therapeutics, in order to enhance productivity, efficiency and / or efficacy.
Owner:SISTEMIC SCOTLAND
A method for predicting the functional impact of somatic mutations in cancer
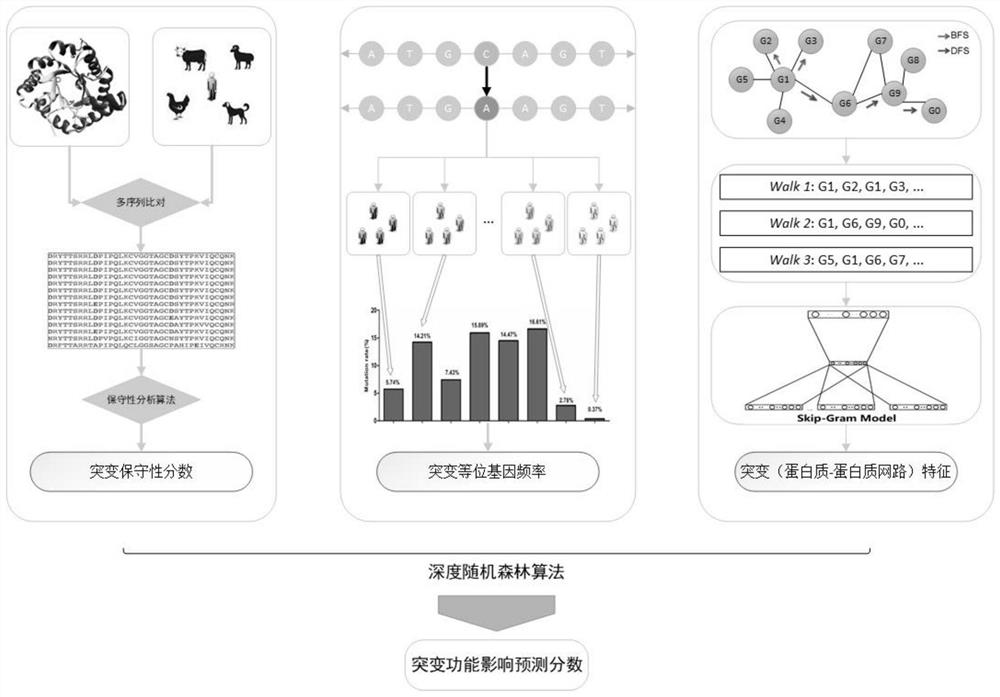
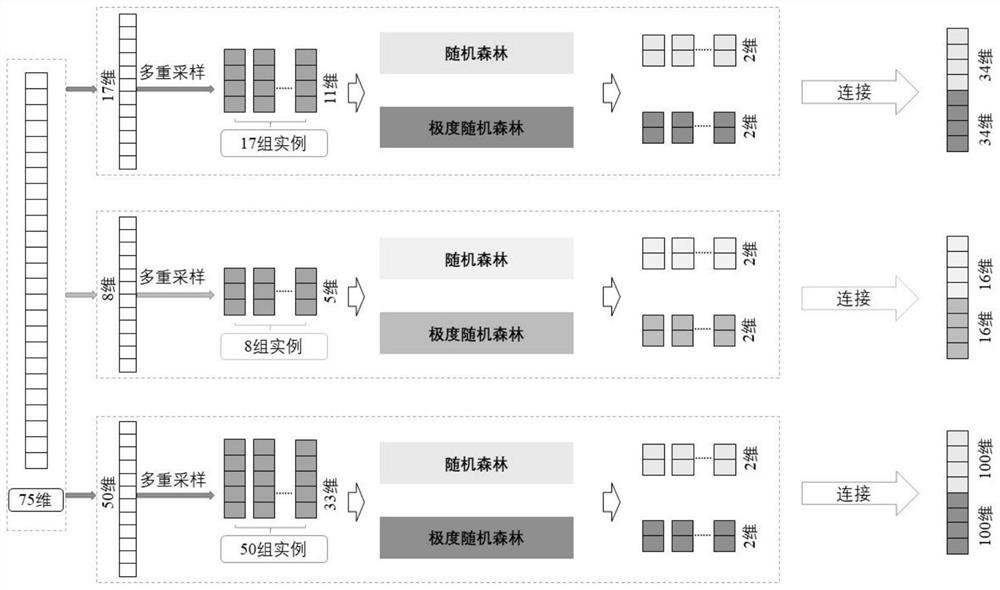
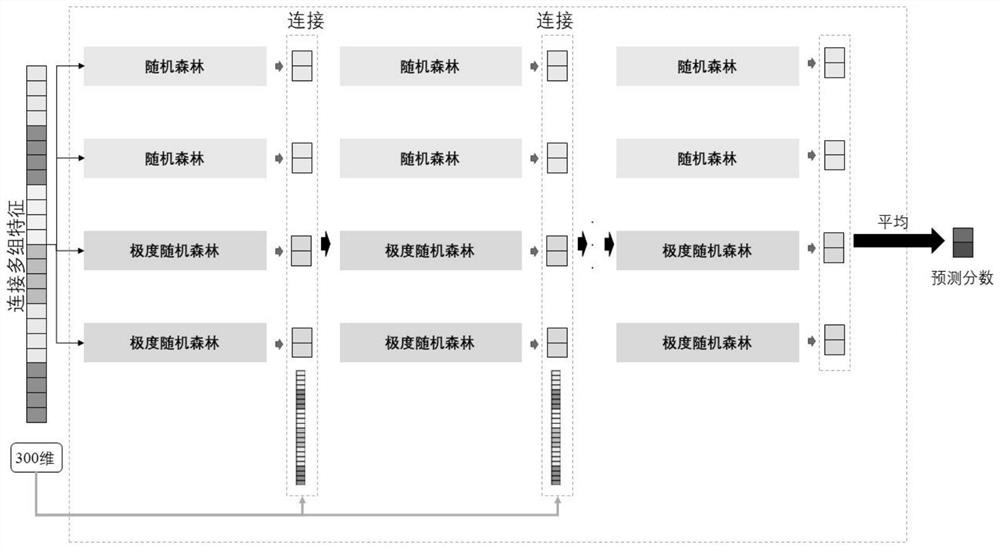
ActiveCN114155910BAccurate predictionEffective predictionBiostatisticsCharacter and pattern recognitionMutation frequencyGene product
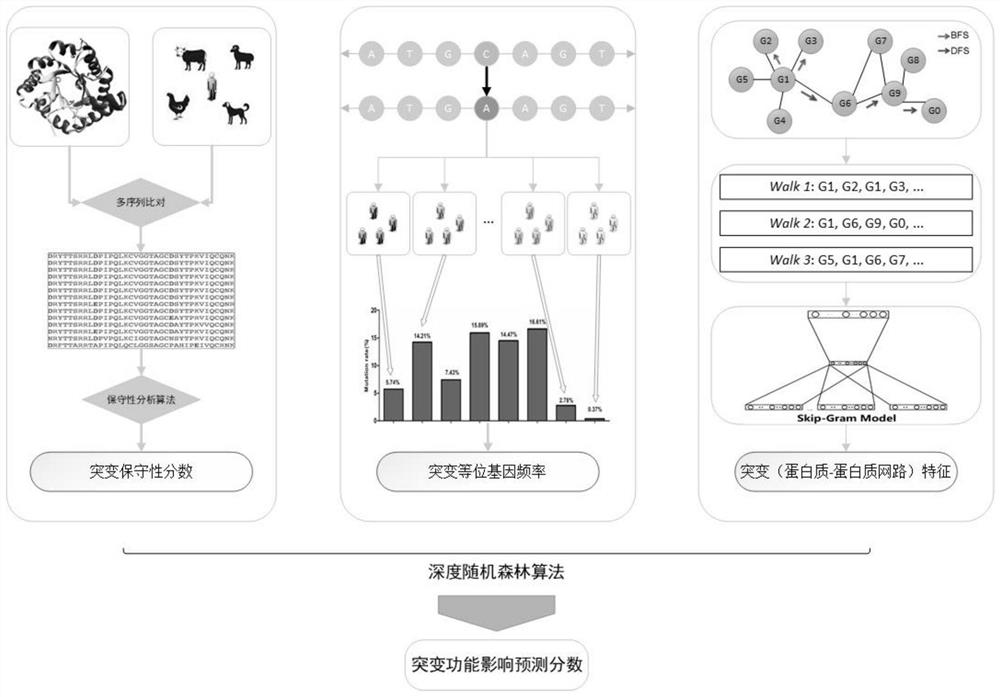
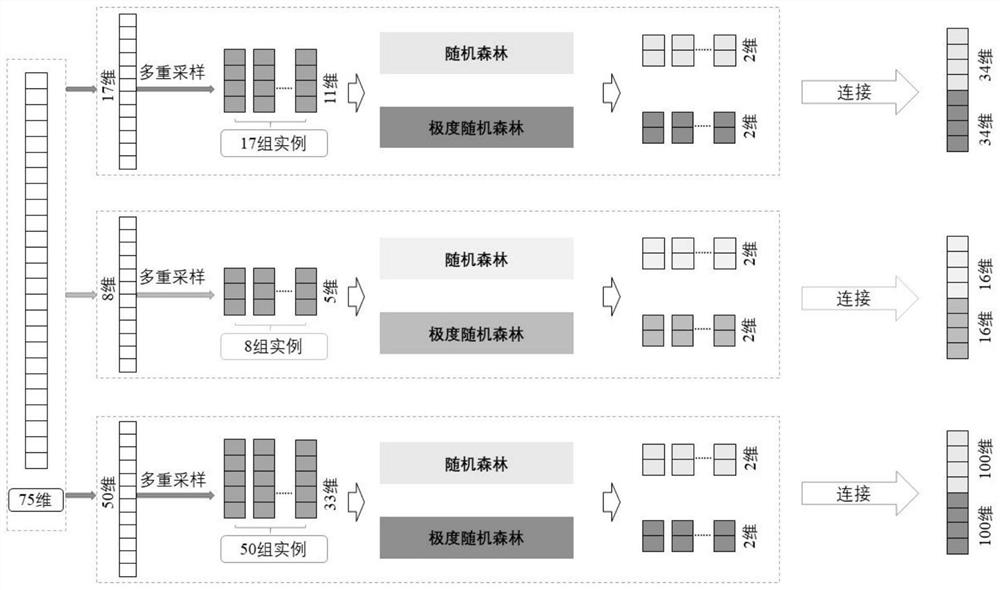
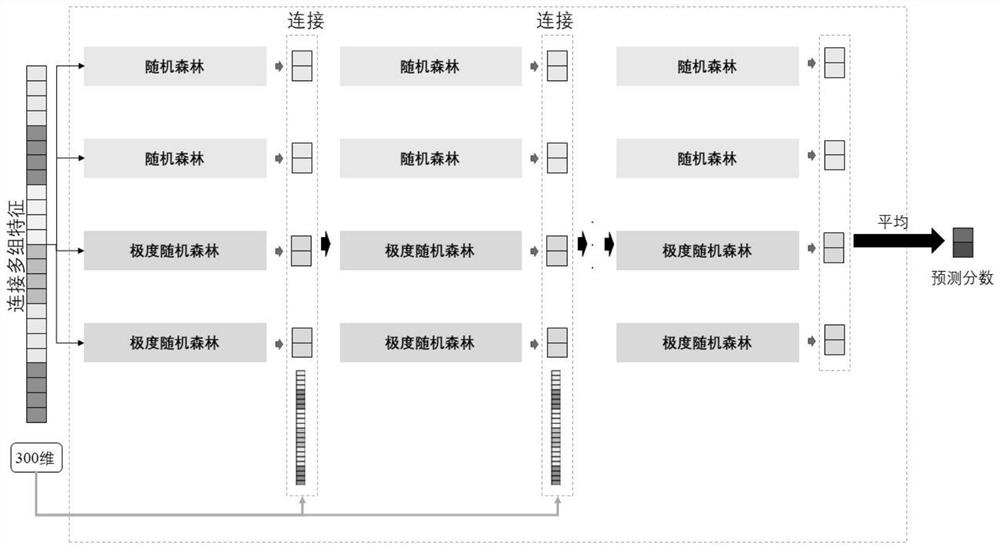
The present invention is a method for predicting the functional impact of cancer somatic mutation. The invention constructs a comprehensive mutation feature set, integrates the mutation frequencies of different populations together; performs conservative estimation based on the mutation sites of multiple species, and estimates the probability that these mutations are harmful mutations; and constructs mutation network features based on the interaction of gene products. , complete the construction of mutation network features; build a deep random forest prediction model based on multiple sampling and hierarchical structure, and make functional predictions for cancer somatic mutations.
Owner:HARBIN INST OF TECH
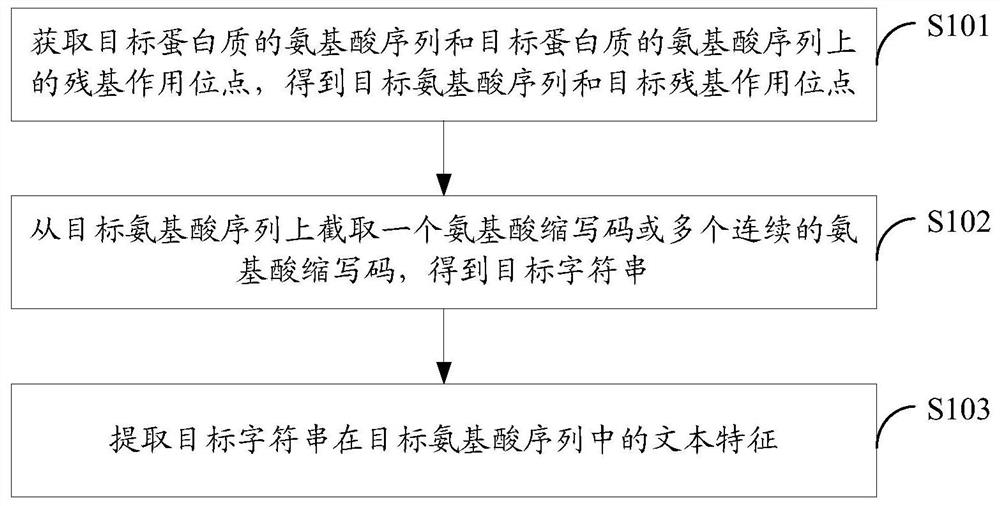
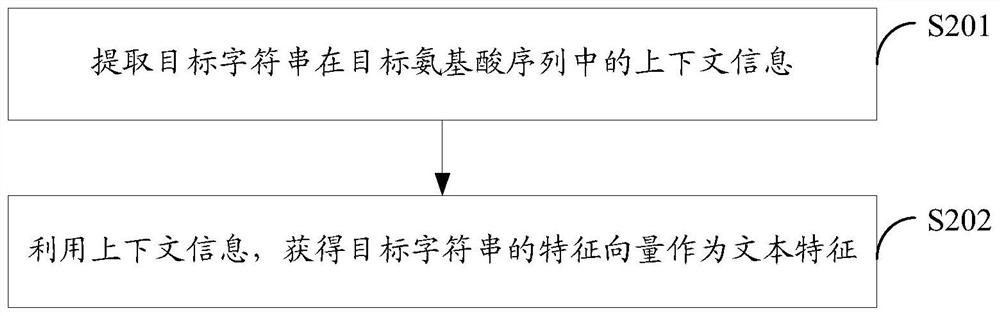
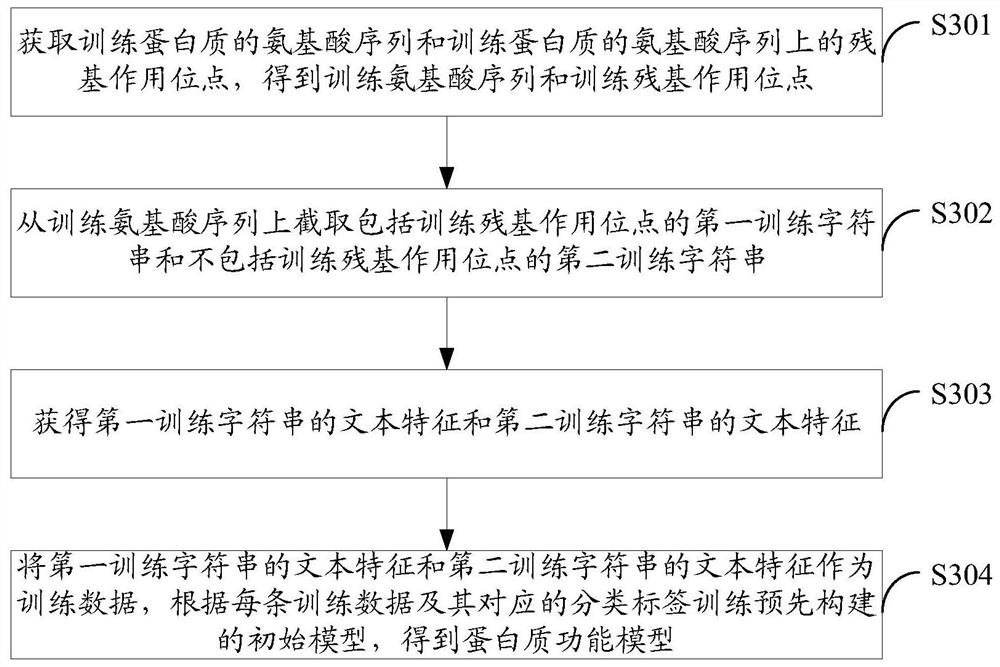
Method and device for protein feature extraction, function model generation, and function prediction
ActiveCN109215737BImprove forecasting efficiencyImprove forecast accuracySequence analysisHybridisationProtein targetProtein function prediction
The embodiment of the present application discloses a method and device for protein feature extraction, function model generation, and function prediction; the protein feature extraction method includes: obtaining the amino acid sequence of the target protein and the residue interaction site on the amino acid sequence of the target protein, Obtain the target amino acid sequence and target residue interaction site; intercept an amino acid abbreviation code or multiple consecutive amino acid abbreviation codes from the target amino acid sequence to obtain a target string including the target residue interaction site; extract the target string in the target The text features in the amino acid sequence, using the feature of the residue interaction site on the amino acid sequence as the expression of protein function, can provide high-quality feature input for the subsequent use of machine learning algorithms for protein function prediction, which is conducive to abandoning time-consuming The powerful manual prediction method improves the efficiency and accuracy of protein function prediction.
Owner:NEUSOFT CORP
Biological material function prediction and evaluation method
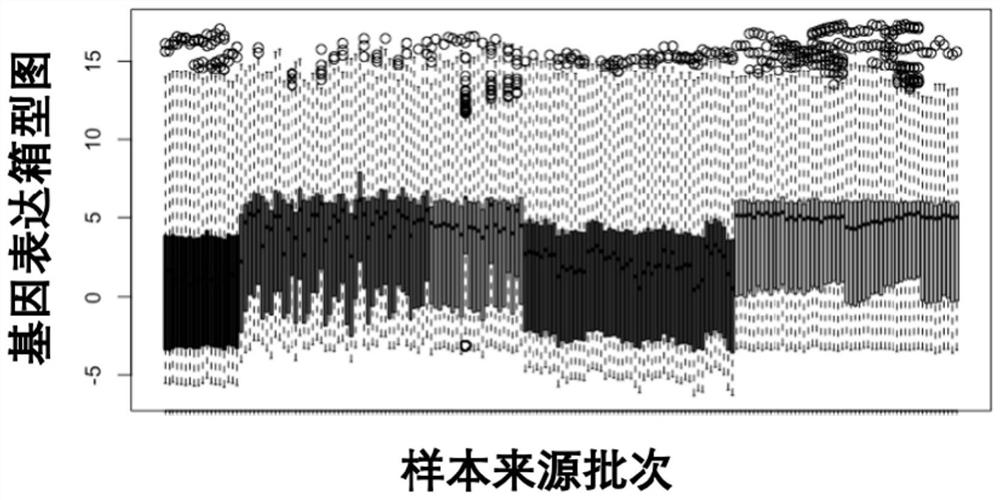
ActiveCN113604544AFull picture of differentiation statusImprove the accuracy of judgmentMicrobiological testing/measurementMachine learningTotal rnaBiological materials
The invention relates to a biological material function prediction and evaluation method, and solves the technical problems of intensive labor, long experimental period and high heterogeneity of a sample library in the existing evaluation method. The biological material function prediction and evaluation method comprises the following steps of (1) culturing human bone marrow mesenchymal stem cells in an environment of a material to be detected; (2) collecting the human bone marrow mesenchymal stem cells cultured in the step (1), extracting total RNA (Ribonucleic Acid), purifying and establishing a library, and sequencing a transcriptome to obtain transcriptome data of a to-be-detected sample; and (3) performing batch effect correction and feature extraction on the transcriptome data of the to-be-detected sample obtained in the step (2), inputting the transcriptome data into a function prediction evaluation model, and calculating the confidence of the to-be-detected sample for different cell types. The method can be used in the field of biological material function prediction and evaluation.
Owner:PEKING UNIV SCHOOL OF STOMATOLOGY
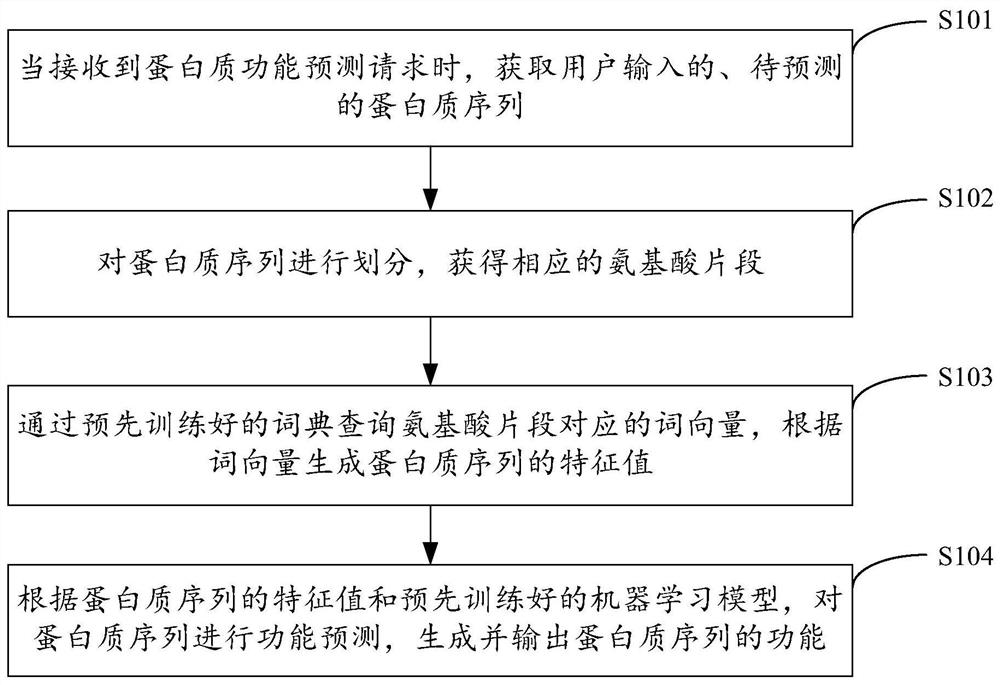
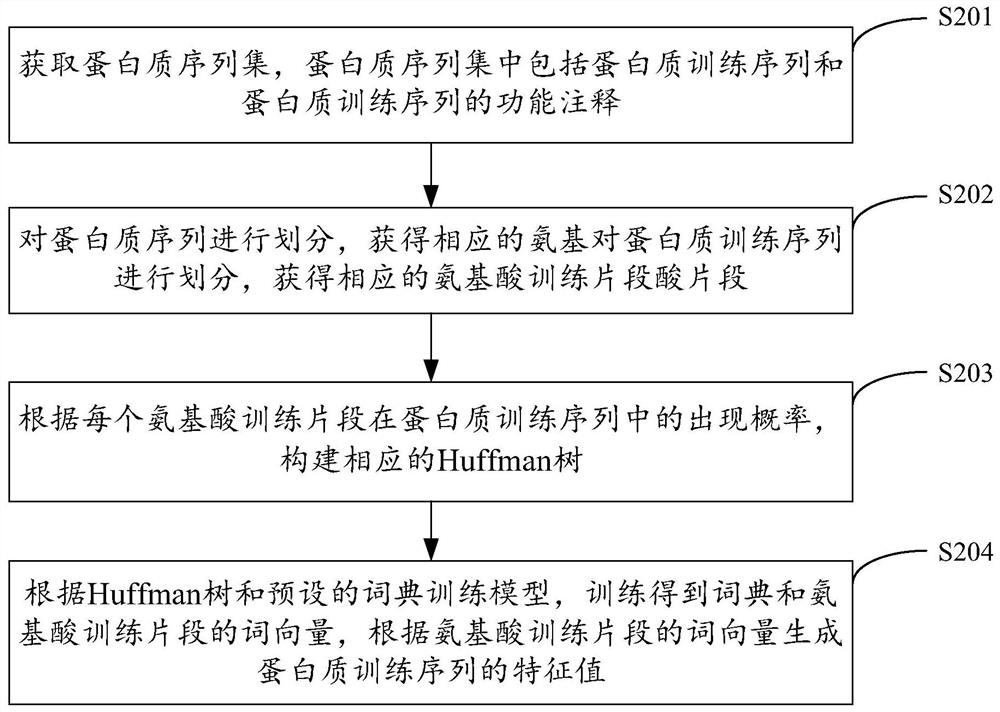
Protein function prediction method, device, equipment and storage medium
ActiveCN109147868BImprove accuracyImprove efficiencyProteomicsGenomicsESA ProteinProtein function prediction
The present invention is applicable to the technical field of biological information, and provides a protein function prediction method, device, equipment, and storage medium. The method includes: obtaining a protein sequence to be predicted, dividing the protein sequence, obtaining corresponding amino acid fragments, and training Query the word vectors corresponding to the amino acid fragments in a good dictionary, generate the eigenvalues of the protein sequence based on these word vectors, predict the function of the protein sequence according to the eigenvalues of the protein sequence and the trained machine learning model, and generate and output the eigenvalues of the protein sequence Therefore, by obtaining the eigenvalues of protein sequences with context characteristics and performing machine learning on these eigenvalues, the accuracy and efficiency of protein function prediction are effectively improved, and the effect of protein function prediction is improved.
Owner:SHENZHEN UNIV
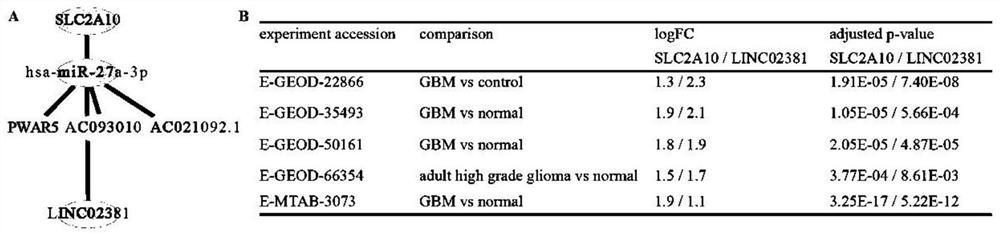
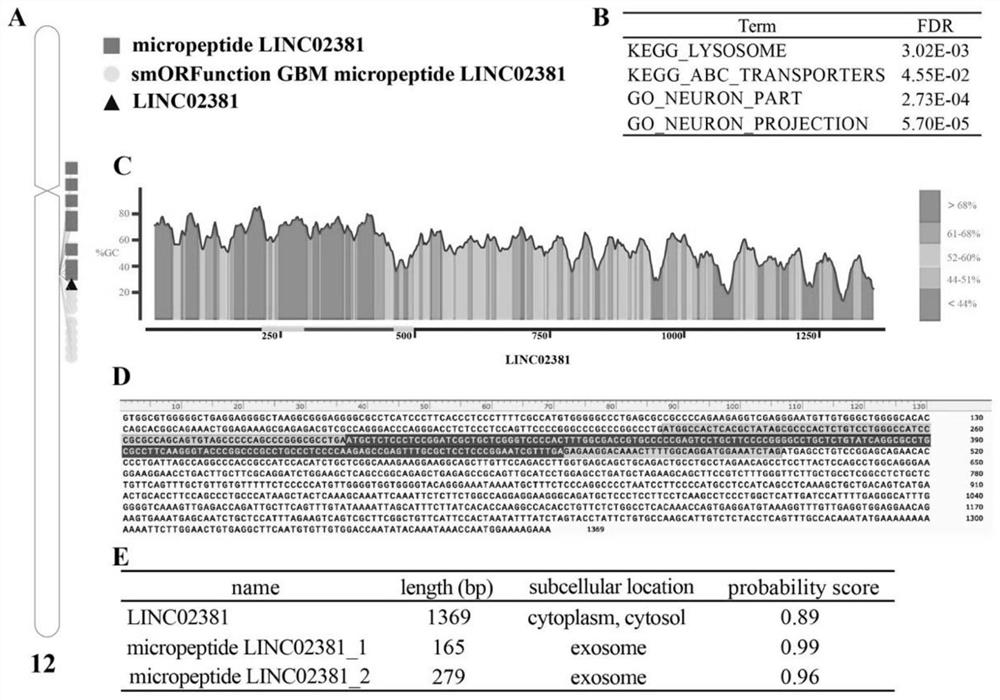
Function prediction and screening method of glioblastoma lncRNA coding micropeptide
The invention discloses a function prediction and screening method of glioblastoma lncRNA coding micropeptides. The method comprises the following steps: screening a glucose transporter gene set; carrying out significant differential expression and survival analysis on the glucose transporter gene set in the pan cancer; verifying the SLC2A10 in the glioblastoma, and predicting ceRNA (Ribonucleic Acid); the method comprises the following steps: carrying out LINC02381 coding to predict the micro-peptide, positioning the predicted micro-peptide into a human chromosome, carrying out identification and function enrichment analysis on the predicted micro-peptide, and reserving the micro-peptide which has the same chromosome as the LINC02381 and is predicted to have potential functions. The invention provides a function prediction and screening method of glioblastoma lncRNA (long non-coding RNA) coded micropeptide, and provides a basis for researching the effect of the lncRNA coded micropeptide in glioblastoma in prognosis or diagnosis.
Owner:THE FIRST AFFILIATED HOSPITAL OF WANNAN MEDICAL COLLEGE YIJISHAN HOSPITAL OF WANNAN MEDICAL COLLEGE
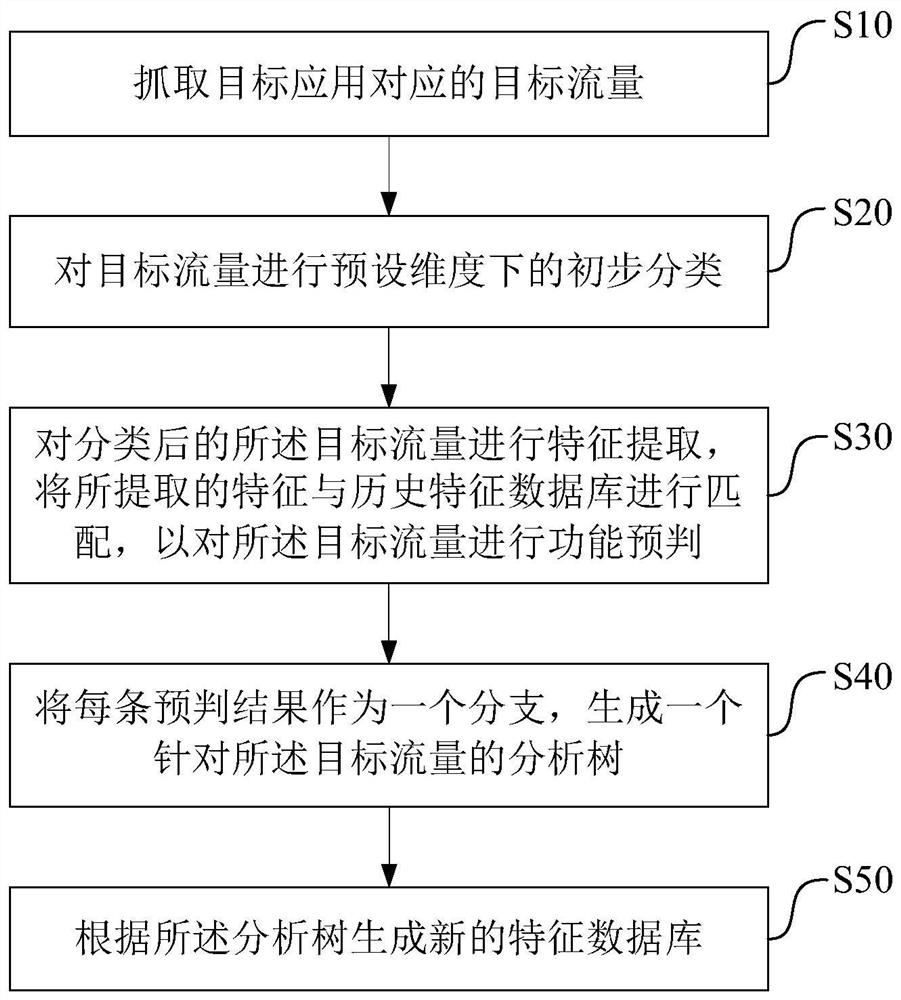
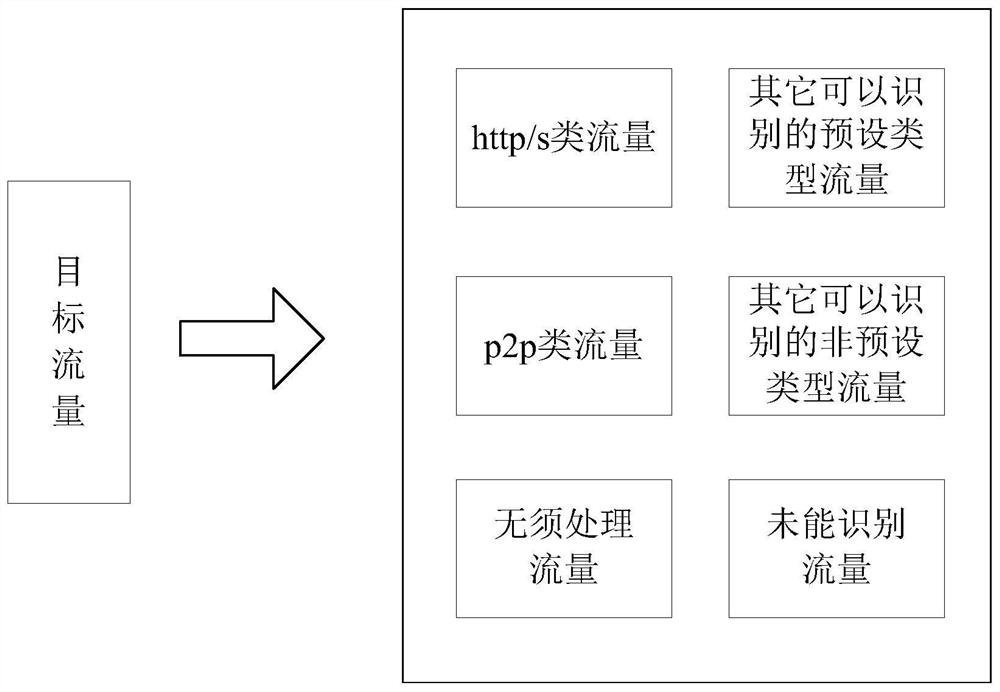
Feature database generation method and device and corresponding traffic sorting method and device
The invention discloses a method for generating a feature database. The method includes: grabbing target traffic corresponding to a target application; performing preliminary classification on the target traffic in a preset dimension; performing feature extraction on the classified target traffic, Match the extracted features with the saved historical feature database to predict the function of the target traffic; use each prediction result as a branch to generate an analysis tree for the target traffic; according to the Parse the tree to generate a new feature database. The present invention also provides a feature database generation device and a traffic sorting method and device using the feature database. The feature database generation method and device, traffic sorting method and device provided by the present invention can perform feature matching during traffic sorting according to the feature database, and optimize traffic matching accuracy and efficiency.
Owner:CHINANETCENT TECH
A functional prediction method for single-nucleotide genomic variation in non-coding regions
ActiveCN109033751BLow sequencing depth requirementAvoid time costProteomicsGenomicsBinding siteNucleotide
The invention discloses a method for predicting the function of a single nucleotide genome variation in a non-coding region, comprising the following steps: 1) identification of an open chromatin region; 2) identification of a transcription factor binding site; 3) evaluation of a single nucleotide variation Role: Based on the site-specific frequency matrix of transcription factors, calculate the impact of single nucleotide variations located in the binding site region of transcription factors on the binding of transcription factors, and identify single nucleotide variations that significantly change the binding ability of transcription factors; further Assess the effect of single nucleotide variants by looking at the biological pathways of transcription factors' target genes. This method uses chromatin open region information and gene expression information to complete the identification of multiple transcription factors and their binding sites at one time, and realize the functional annotation of genomic variation in non-coding regions.
Owner:SOUTHEAST UNIV
Cancer somatic mutation function influence prediction method
ActiveCN114155910AAccurate predictionEffective predictionBiostatisticsCharacter and pattern recognitionMutation frequencyGene product
The invention relates to a cancer somatic mutation function influence prediction method. According to the method, a comprehensive mutation feature set is constructed, and mutation frequencies of different populations are integrated together; carrying out conservative estimation based on mutation sites of multiple species, and estimating the probability that the mutations are harmful mutations; mutation network feature construction based on gene product interaction is carried out, and mutation network feature construction is completed; and constructing a deep random forest prediction model based on multiple sampling and a hierarchical structure, and performing function prediction on cancer somatic mutation.
Owner:HARBIN INST OF TECH
Corn variable splicing isomer function prediction system based on tissue specificity
PendingCN114863992ATake advantage ofImprove function prediction effectBiostatisticsSystems biologyNonnegative matrix factorisationGenetics
The invention provides a corn variable splicing isomer function prediction system based on tissue specificity, which is characterized in that isoform co-expression networks of a plurality of tissues are constructed by processing expression data of isoform on each tissue, and the isoform co-expression networks of each tissue are integrated through self-adaptive weight, so that the function of the corn variable splicing isomer is predicted. A high-quality isoform organization specific association network can be obtained; according to the method, an isoform sequence similarity network is constructed by using isoform sequence data, and the isoform sequence similarity network is fused with an isoform organization specificity association network, so that a better isoform function association network can be obtained; multi-example learning is performed through non-negative matrix factorization, and meanwhile, the non-negative matrix factorization is guided by using an isoform function association network, so that more accurate and more comprehensive function prediction of isoform is realized.
Owner:SHANDONG UNIV
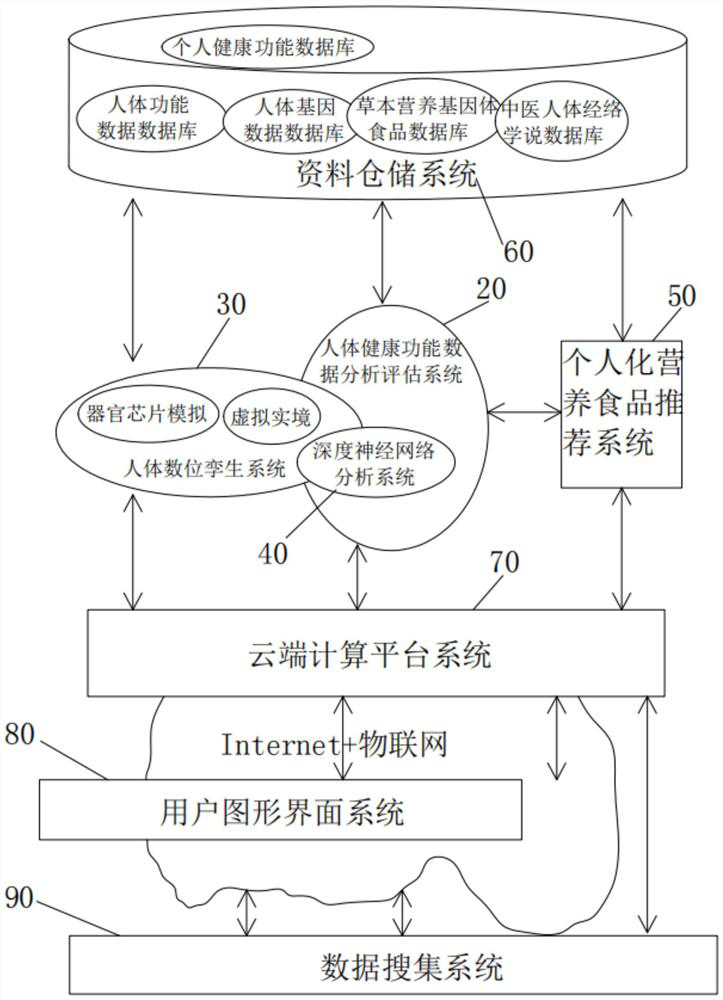
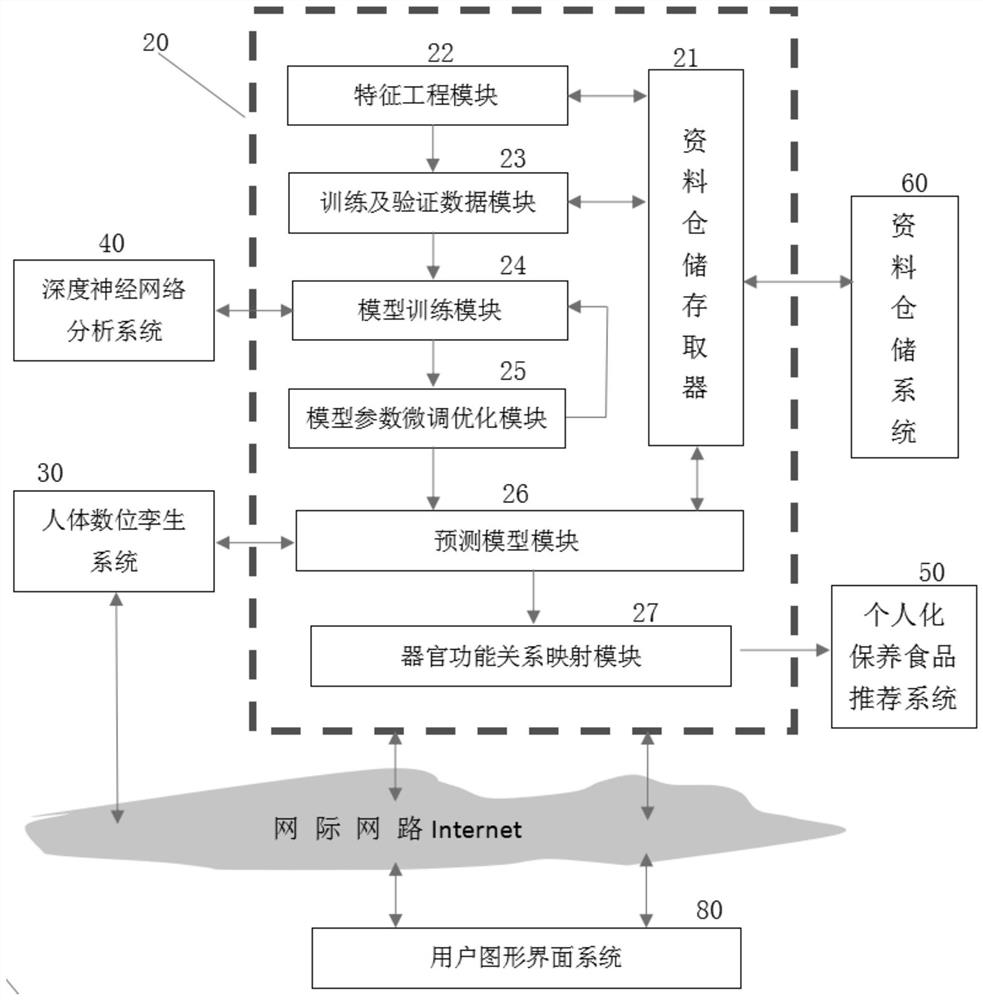
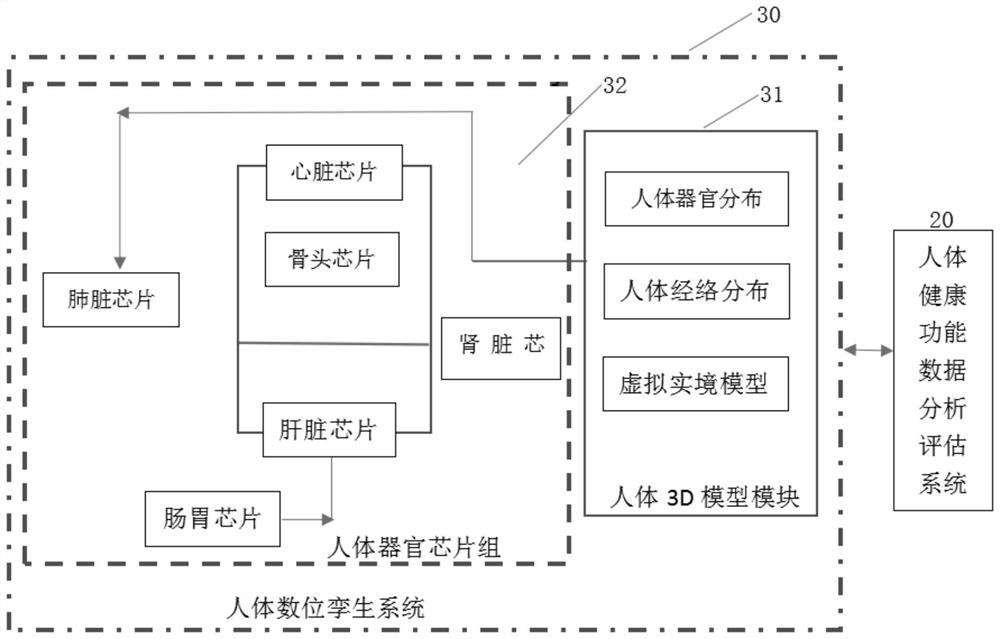
Computer-aided system and method for preventive maintenance of human health in life cycle
The invention discloses a computer-aided system and method for human health life cycle preventive maintenance, and relates to the technical field of artificial intelligence. Comprising a human body health function data analysis and evaluation system, a human body digital twin system, a deep neural network analysis system, a human nutrition food recommendation system, a data storage system, a cloud computing platform system, a user graphical interface system and a data collection system. Whether the human body has potential diseases or diseases can be predicted, the human body function is analyzed according to the data in the back-end data warehouse, whether the human body has potential diseases or diseases or not is predicted, warning and suggestions are provided for preventive measures, and the purpose of preventive maintenance of the human body health life cycle is achieved.
Owner:益波信息科技(上海)有限公司
Novel coronavirus sample base mutation automatic assessment method
PendingCN113936739AComprehensive assessmentAccurate judgmentBiostatisticsProteomicsOriginal dataMedicine
The invention discloses a novel coronavirus sample base mutation automatic assessment method, medium and equipment, and the method comprises the steps: obtaining original data of a novel coronavirus sample to be assessed, and carrying out the comparison to generate an intermediate file; analyzing the intermediate file to obtain base sequence information, and comparing the base sequence information of the novel coronavirus sample to be evaluated with a reference sequence to obtain mutation information; classifying the novel coronavirus sample to be assessed according to the mutation information to generate classification information; performing epidemic analysis, function prediction and clinical association on the novel coronavirus sample to be assessed, and generating assessment data according to an epidemic analysis result, a function prediction result, a clinical association result and the classification information; according to the method, the novel coronavirus sample base mutation can be automatically and comprehensively assessed so that the virus sample is accurately judged, and powerful help is provided for virus traceability, virus typing and virus research.
Owner:SICHUAN UNIV
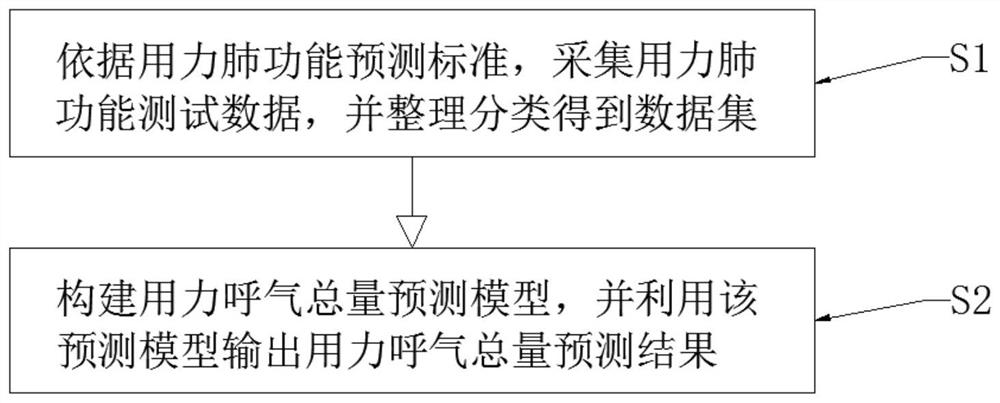
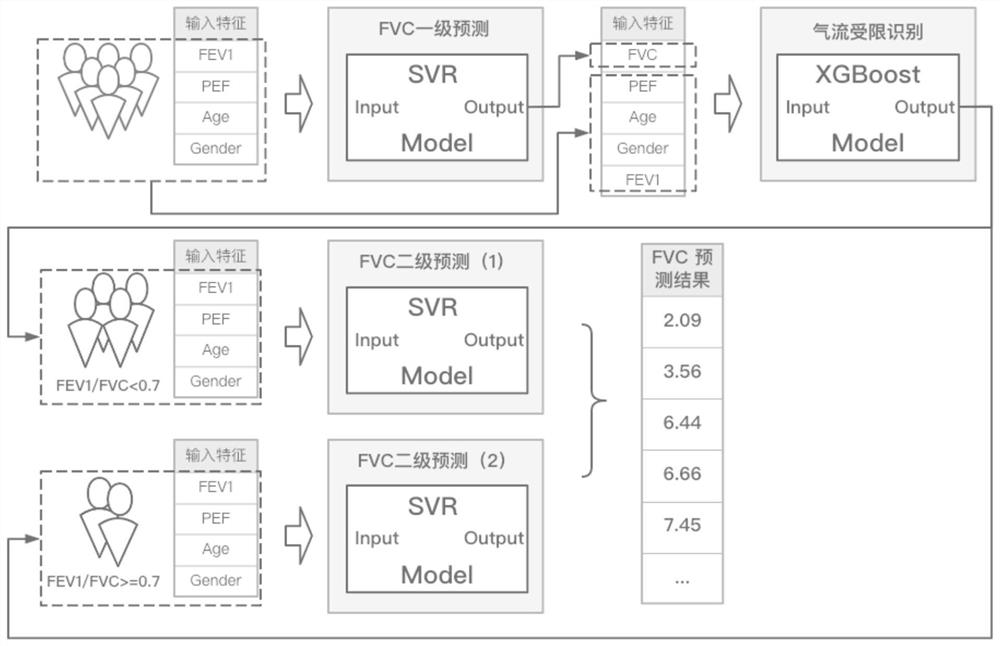
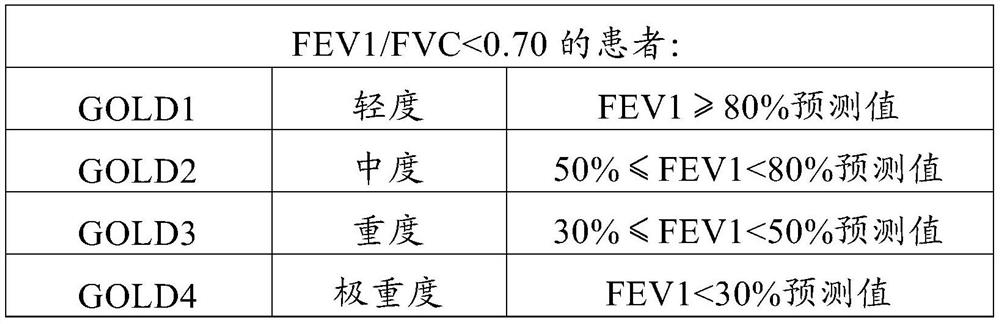
Forced expiration total amount prediction method
PendingCN113469227ASolve the resultAddress issues that affect its diagnostic effectivenessCharacter and pattern recognitionDiagnostic recording/measuringData setForced expiration
The invention discloses a forced expiration total amount prediction method, which comprises the following steps: S1, according to a forced lung function prediction standard, collecting forced lung function test data, and sorting and classifying the data to obtain a data set; and S2, constructing a forced expiration total amount prediction model, and outputting a forced expiration total amount prediction result by using the prediction model. The method has the beneficial effects that by providing the forced expiration total amount prediction method, a tester can still obtain an FVC value under the condition that the whole test is not completed, so that the problem that the diagnosis effect is influenced due to the fact that an FVC detection result does not meet the acceptability because it is difficult for the tester to insist in expiration time long enough is effectively solved.
Owner:RES INST OF NANJING RUNNAN MEDICAL ELECTRONICS CO LTD +1
Soybean bi-1 gene and its application
ActiveCN108220303BPositive regulation of symbiotic nodulationImprove nitrogen fixation capacityBacteriaMicroorganism based processesBiotechnologyCDNA library
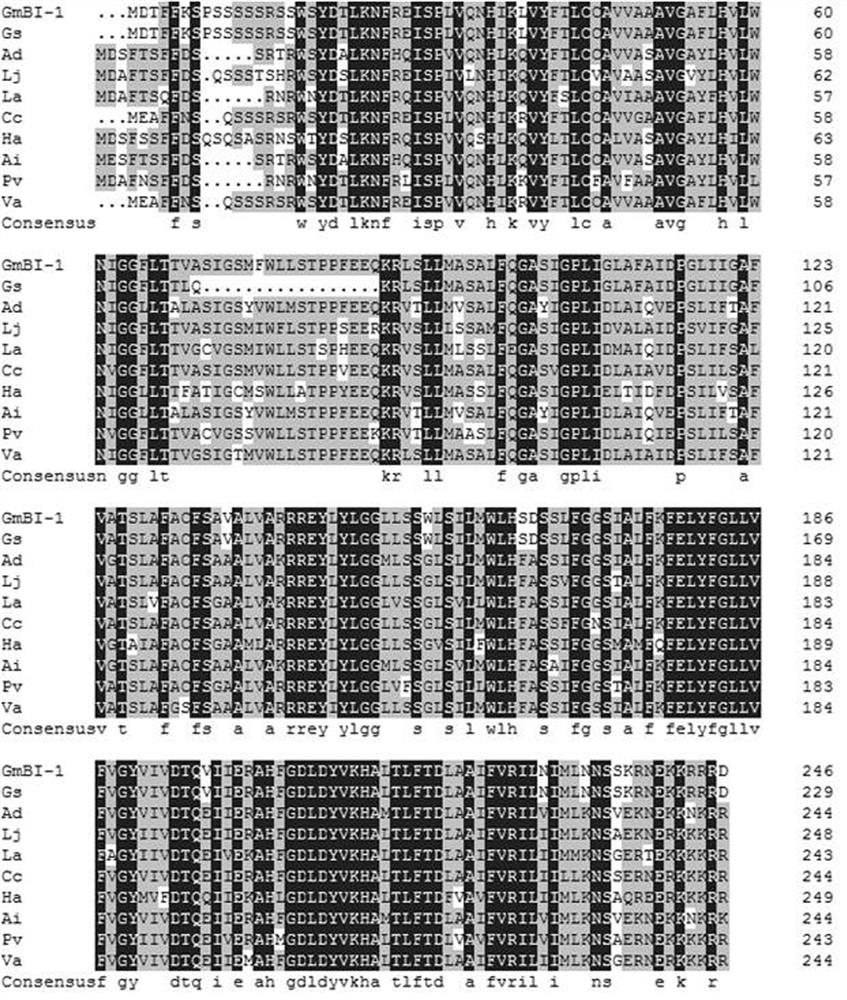
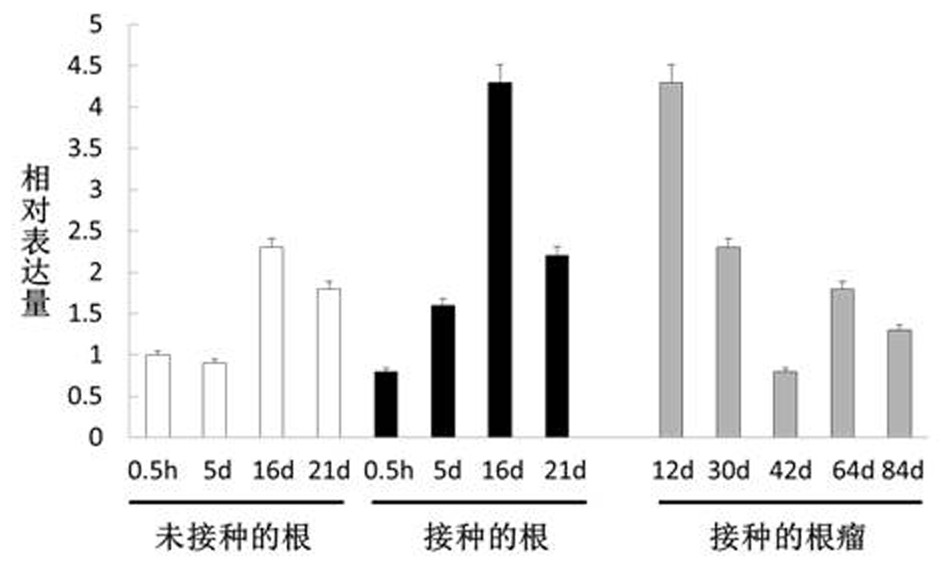
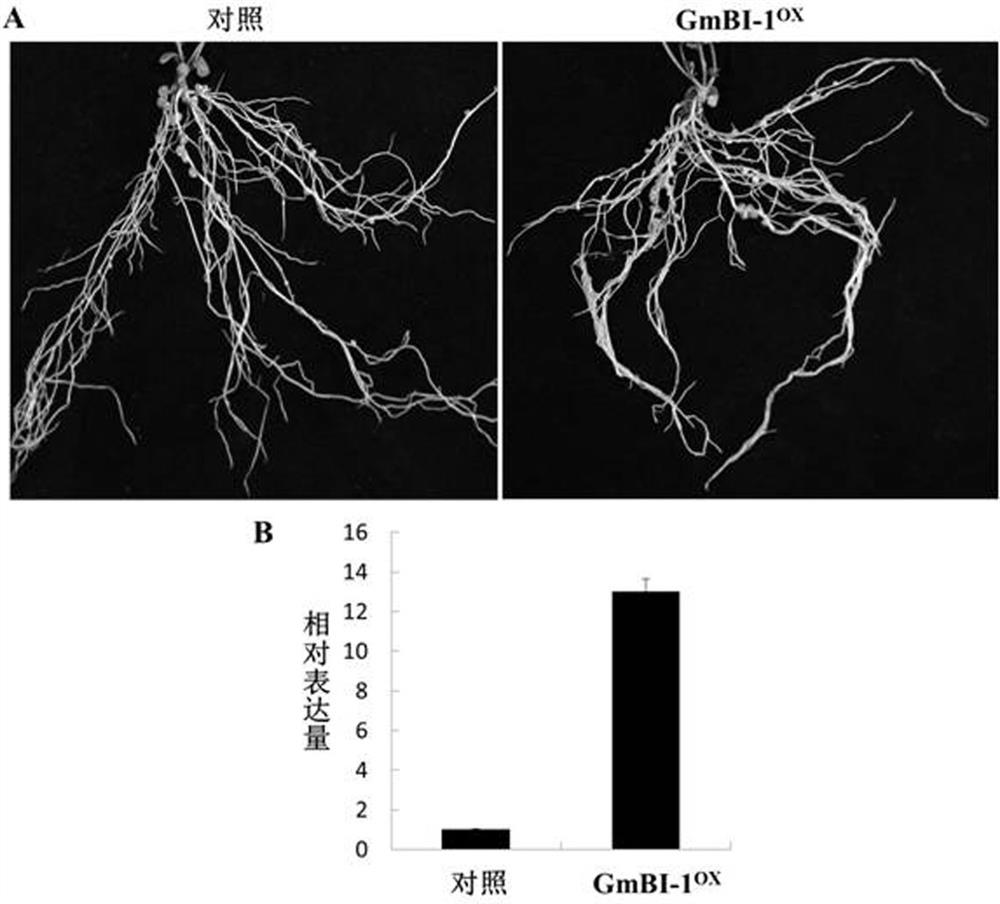
The invention discloses a gene BI-1 of cell withering inhibition factors for regulating and controlling symbiotic nodulation of leguminous plants, and provides application of the gene in regulating and controlling number of root nodules of leguminous plant. A nucleotide sequence of the gene is shown in SEQ ID NO:1 in a sequence list. The gene has the advantages that a yeast two-hybrid technique isutilized to screen in a soybean root nodule AD-cDNA library, a kind of new protein which interacts with receptor protein of nodulation factor is found, and the gene is proofed to be a cell apoptosisinhibition factor gene through methods of sequence comparison, function prediction and the like; an over-expression technique is utilized to study the function of the gene in the symbiotic nodulationprocess of crowtoe of the leguminous plant; proofed by results, the protein encoded by the gene participates in the symbiotic nodulation process, the number of the root nodules is obviously increasedafter over-expression, and the application prospect on the symbiotic nitrogen immobilizing of the leguminous plant is realized.
Owner:XINYANG NORMAL UNIVERSITY
Methods and systems for determination of an effective therapeutic regimen and drug discovery
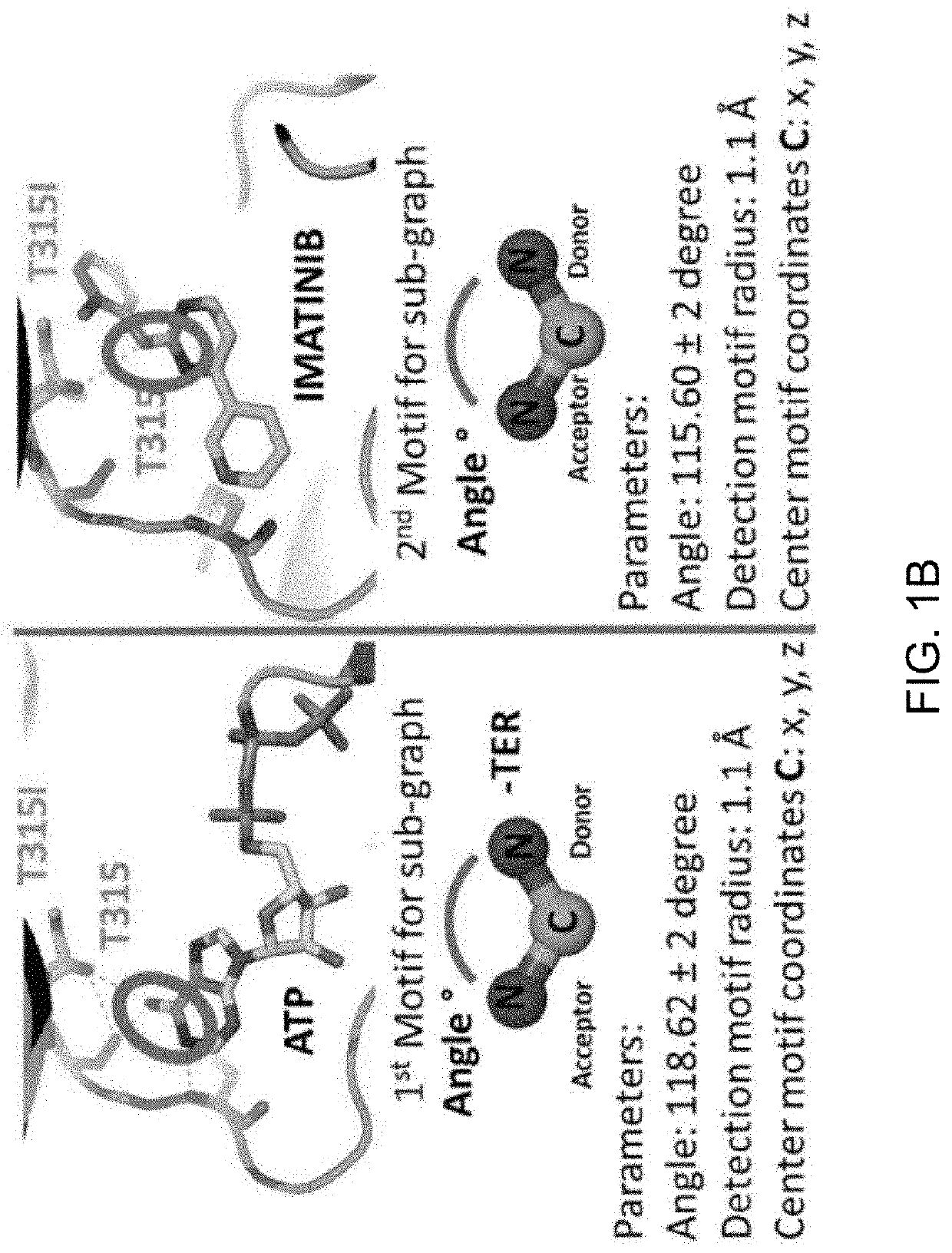
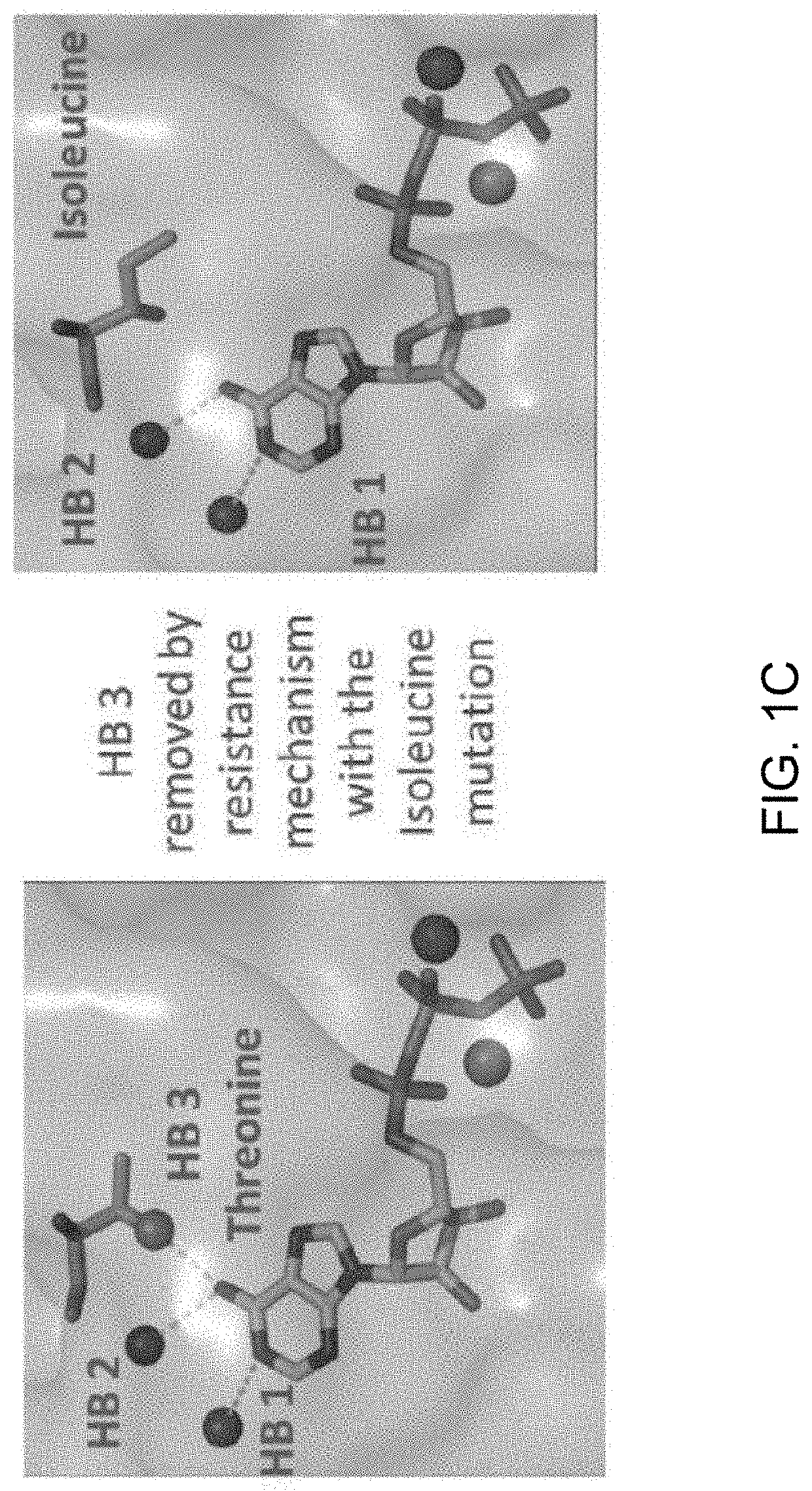
ActiveUS20200208222A1Chemical property predictionMicrobiological testing/measurementGenes mutationActivating mutation
The present invention relates to the discovery of a method for identifying a treatment regimen for a patient diagnosed with cancer, predicting patient resistance to therapeutic agents and identifying new therapeutic agents. Specifically, the present invention relates to the use of an algorithm to identify a mutation in a kinase, determine if the mutation is an activation or resistance mutation and then to suggest an appropriate therapeutic regimen. The invention also relates to the use of a pattern matching algorithm and a crystal structure library to predict the functionality of a gene mutation, predict the specificity of small molecule kinase inhibitors and for the identification of new therapeutic agents.
Owner:DNA SEQ 2 INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com