Cancer somatic mutation function influence prediction method
A technology of somatic mutation and prediction method, applied in the field of prediction of the impact of somatic mutation of cancer, can solve the problem of general prediction effect, and achieve the effect of predicting the effect of mutation function
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 1
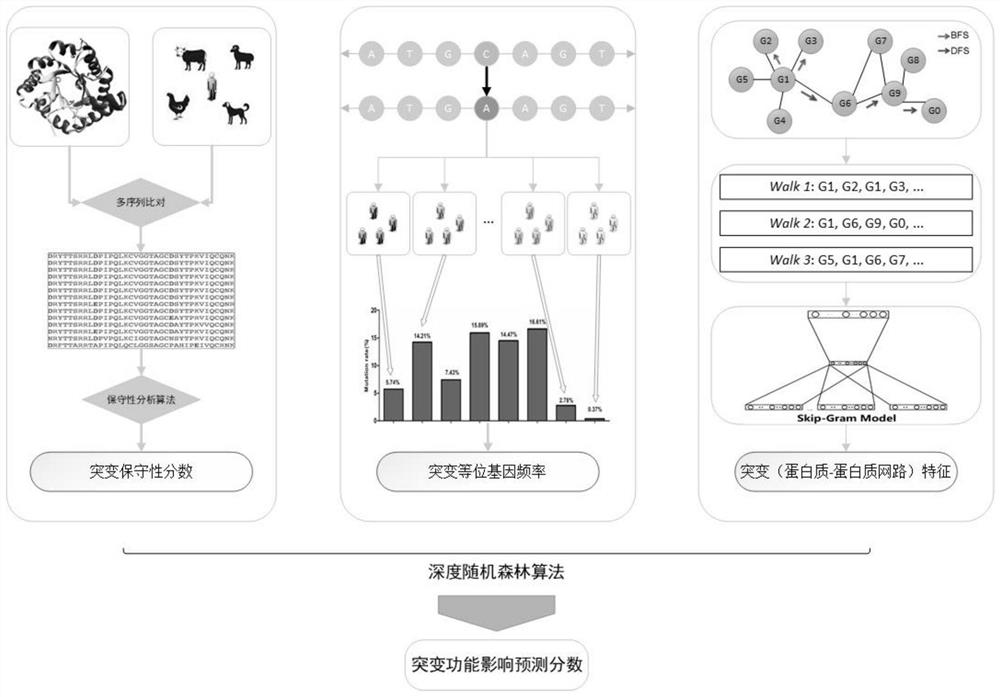
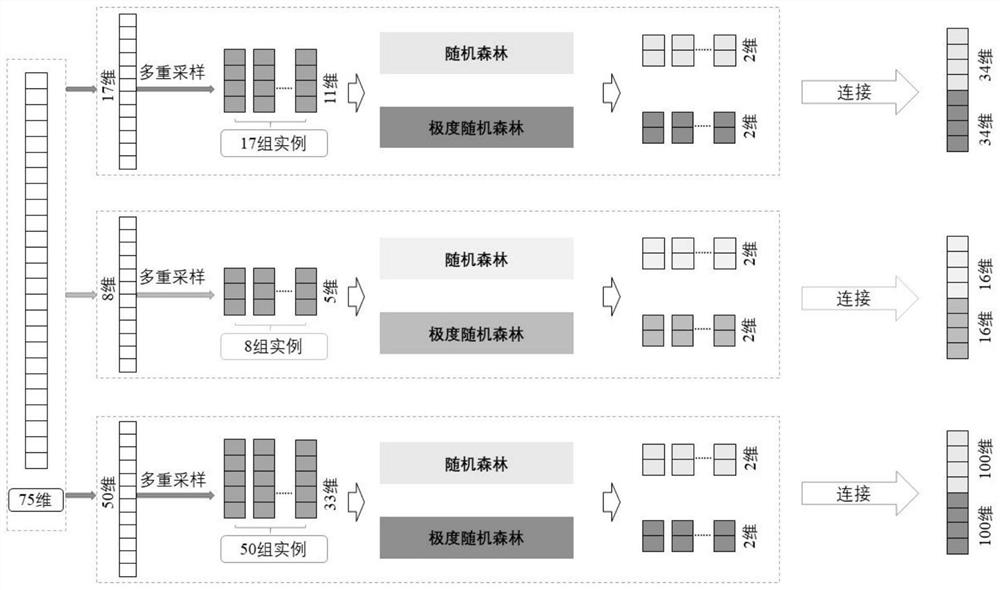
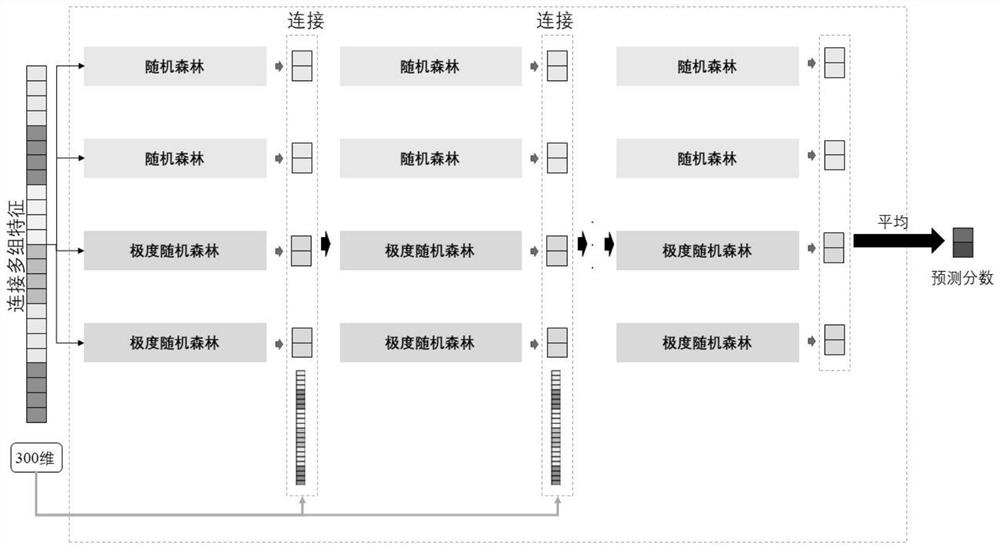
[0036] according to Figure 1 to Figure 4 As shown, the specific optimized technical solution adopted by the present invention to solve the above technical problems is: a method for predicting the functional impact of cancer somatic mutations, comprising the following steps:
[0037] A method for predicting the functional impact of cancer somatic mutations, comprising the following steps:
[0038] Step 1: Construct a comprehensive mutation feature set to integrate the mutation frequencies of different populations together;
[0039] The step 1 is specifically:
[0040] Mutations have different frequencies in different populations. When some mutations have a higher frequency in the population, they tend to be harmless mutations, and when some mutations occur at a lower frequency, they tend to be harmful mutations; the mutation frequencies of different populations Integrate together as a class of predictive features to predict the functional impact of mutations.
[0041] Step ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com