Molecular recognition feature function prediction method based on ensemble learning
A molecular identification and integrated learning technology, applied in the field of bioinformatics, can solve the problems of high resource consumption, complex experimental process and high cost, and achieve the effect of low resource consumption, simple experimental process and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] The specific embodiments and effects of the present invention will be further described in detail below in conjunction with the accompanying drawings.
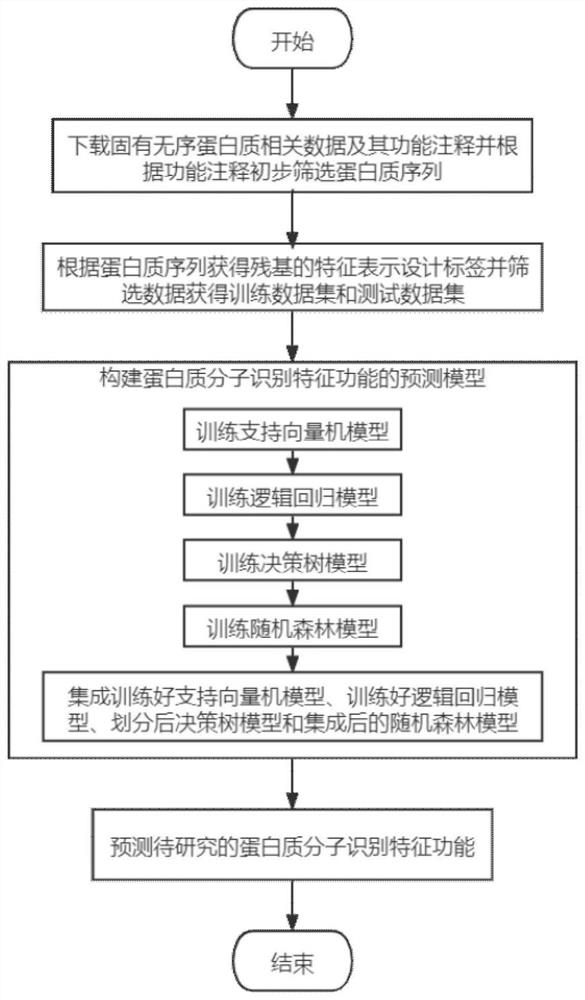
[0026] refer to figure 1 , the implementation steps of this example are as follows:
[0027] Step 1. Obtain data related to inherently disordered proteins and their functional annotations, and preliminarily screen protein sequences based on functional annotations.
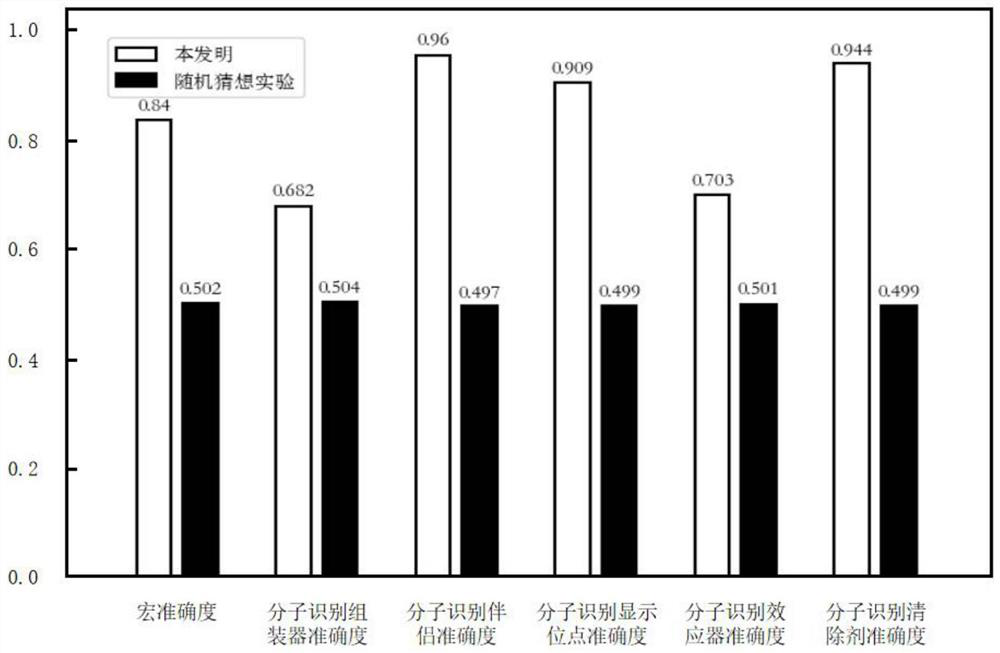
[0028] 1.1) Download the 2020_12 version of the data set in the DisProt database from the public website, including 1590 inherently disordered protein sequences and corresponding 7 functional annotations. The 7 functional annotations are entropy chain, bioconcentration, molecular recognition assembler, molecular Recognition partner, molecular recognition display site, molecular recognition effector and molecular recognition scavenger, in which molecular recognition assembler, molecular recognition partner, molecular recognition display site, molecular recog...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com